1 Introduction
Synthetic oligonucleotides can be potentially used for the specific inhibition of gene expression. Thus, they represent an attractive therapeutic approach for the treatment of various viral diseases. These oligonucleotides can cause the inhibition of gene expression by targeting either the m-RNA (antisense/siRNA mechanism) [1,2] or double stranded DNA (triplex formation) [3] or proteins (aptamer selection) [4]. The effective application of oligonucleotide as a drug is, however, seriously limited due to the poor cellular uptake and targeting of these molecules. The conjugation of oligonucleotides to already known cell penetrating peptide vectors offers an alternative way to overcome the problem of poor uptake. Such peptide–oligonucleotide conjugates (POC) have been shown to enhance the cell specific targeting, uptake efficiency and stability to degradation in comparison to unmodified oligonucleotides. Besides, in many cases, these conjugates show enhanced binding with the target sequence [5].
Chemical synthesis of the POC has therefore generated considerable interest. Different methods for the preparation of POC have been described and these can be classified into two broad approaches [5]. First is the stepwise solid phase synthesis that involves the preparation of the peptide and oligonucleotide fragments on the same solid support. This approach has only a limited application because of the poor compatibility between the oligonucleotide and peptide chemistries. Second is the fragment coupling approach that involves separate solid phase assembly of the oligonucleotide and peptide fragments followed by the solution phase coupling of the two deprotected fragments. This can be accomplished by introducing mutually reactive groups into each fragment during the solid phase synthesis. The coupling leads to the formation of chemical linkages like disulphide, thioether, amide or maleimide [5]. The fragment coupling approach has found wider acceptability on account of excellent coupling efficiencies and ease of purification. This also offers the possibility of coupling almost any peptide to the oligonucleotide. Moreover, the process can be generalised to anchor various other reporters such as carbohydrates, fluorophores and so on to the oligonucleotides.
The earlier work from our laboratory has focussed on the use of chemoselective oxime ligation for efficient conjugation of oligonucleotide to variety of molecules such as peptides, carbohydrates and fluorescent reporter groups. It has been shown that oxime bond formation can be successfully employed to prepare the oligonucleotide conjugates bearing peptides at either 5′ or 3′ ends of the oligonucleotide [6,7]. The methodology has been further applied to the labelling of oligonucleotides and RNA [8] and also for anchoring the oligonucleotides on the glass surface [9]. Clearly, the oxime bonds have certain advantages over other chemical linkages that are being employed in the POC synthesis through fragment coupling approach.
In this account, we present a summary of the work accomplished in our laboratory in the area of oligonucleotide conjugation through oxime bond formation. Some of the new problems that are being presently pursued have also been included.
2 Chemoselective oxime ligation
The various methods reported so far, for the synthesis of POCs has been discussed earlier [5]. These can be employed for the synthesis of the POCs but they present certain limitations. For instance, the thiol and/or amine based conjugation lacks regiospecific ligation when reacted with peptides containing multiple cysteines/lysines. One major problem is the possibility of competing reagent hydrolysis along with the cross reactivity with other functional groups. On the other hand, oxime bonds are chemoselective and hence do not require the use of either a protecting group strategy or a preactivation step. The reaction is rapid and usually carried out at slightly acidic pH when the aliphatic amino groups on peptides remains largely protonated, which aids in the solubilisation of the peptides in aqueous solution either alone or with cosolvents. The oxime bond formation could easily be achieved by the reaction of oligonucleotide bearing aldehyde functionality with the reporters carrying the aminooxy function. There could be two methods to introduce an aldehyde group into the oligonucleotides. First involves the use of protected aldehyde moiety, mainly as acetal. This may not be suitable because acidic deprotection step used for the removal of acetal may lead to depurination. The more convenient second route involves the use of a suitably protected 1,2-diol, from which the aldehyde group can easily be generated by mild periodate oxidation. In the following sections, the strategy for the generation of aldehyde function at various sites of the oligonucleotides and subsequent conjugation through oxime bond is discussed.
2.1 Oxime ligation for base modification (Scheme 1)
The oxime ligation was first used for the fluorescent labelling at C8 position of adenine in an 11-mer oligonucleotide [10]. The reaction was achieved using the oligonucleotide 1, bearing the aldehyde functionality at the preselected position, with the fluorescein derivative 2, bearing the complementary reactive aminooxy function. The aldehyde containing oligonucleotide 1, was prepared by oxidation of the corresponding alkenyl oligonucleotide 3. The cleavage of alkene with osmium tetroxide in acetone/water yielded the intermediate oligonucleotide 4, carrying the 1,2-diol moiety. The aldehyde function was generated by the periodate oxidation of the vicinal diol.
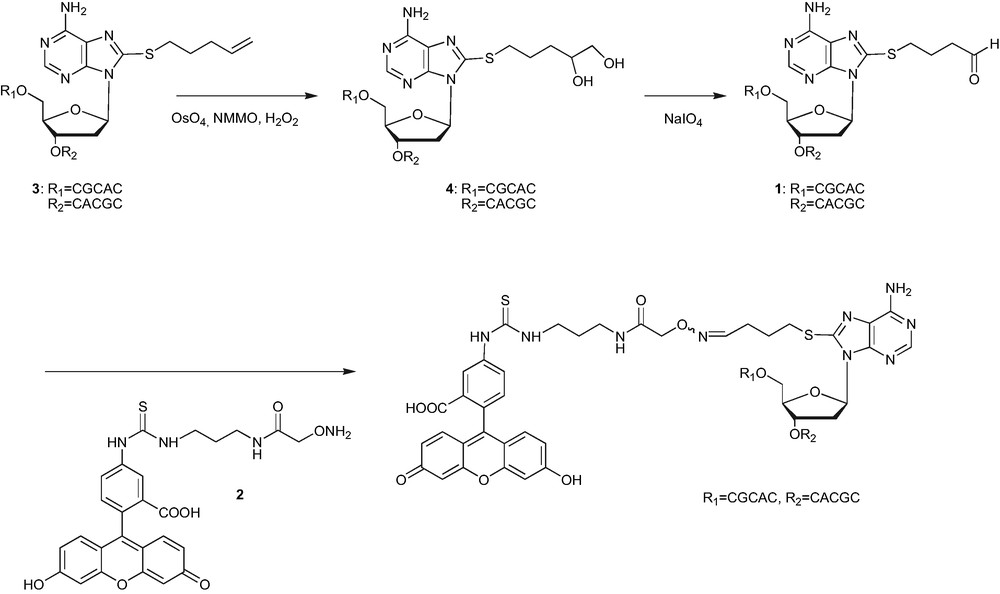
Fluorescent labelling at C8 position using oxime bond strategy.
This result is important in the fact that it shows the usefulness of the process for efficient derivatisation of the oligonucleotides, potentially at any preselected position. Furthermore, the reaction was found to be chemoselective and hence proceeded with exclusive formation of the product. Besides, periodate oxidation did not induce any degradation of the oligonucleotides. We therefore decided to utilise the oxime ligation to prepare different peptide and carbohydrate–oligonucleotide conjugates.
2.2 Oxime ligation for 5′ conjugation (Schemes 2 and 3)
The preparation of the oligonucleotides carrying an aldehyde linker at the 5′ extremity is known [11]. However, the method involves an arduous synthesis of a phosphoramidite linker bearing bis-benzoyl-protected diol in a five-step procedure. We therefore decided to develop a more easy, convenient and straightforward procedure for the synthesis of phosphoramidite linker carrying the protected diol. Consequently, the modified phosphoramidite linkers 6–7 were synthesised. These were prepared in two straight steps starting from 1,2,6-hexane triol and were incorporated at the 5′ end of the oligonucleotide during the solid phase oligonucleotide synthesis using the routine coupling procedure [6a]. Acetic acid treatment afforded the oligonucleotide 8 carrying the 5′ diol moiety. It must be mentioned that the phosphoramidites 6 or 7 being hydrophobic in nature, aids in the oligonucleotide purification during reverse-phase HPLC and the protecting benzylidene group is easily removed by the treatment with 80% acetic acid, conditions routinely employed for the removal of DMTr group in oligonucleotide chemistry. The aldehyde containing oligonucleotide 9 was generated by periodate oxidation.
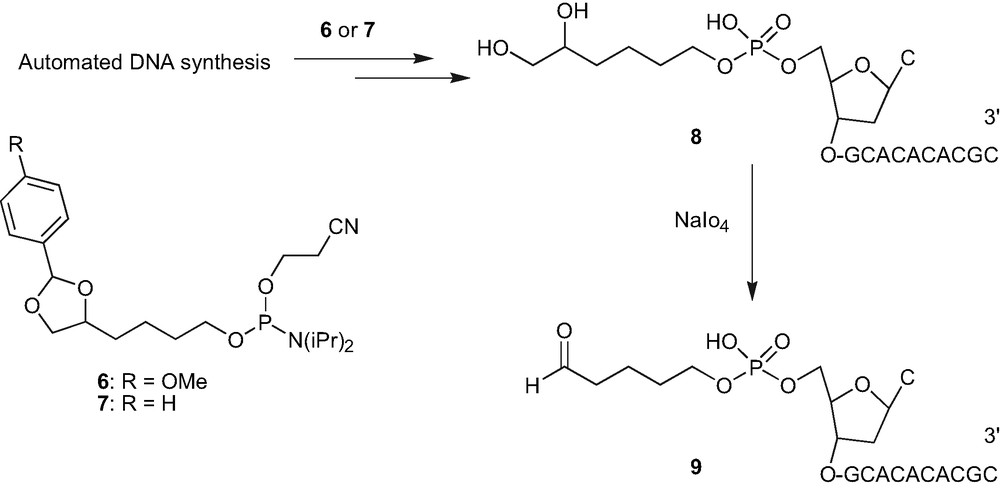
Preparation of 5′ aldehyde containing oligonucleotide 9.

Conjugation at the 5′ extremity via oxime bond formation.
The oligonucleotide 9 bearing the 5′ aldehyde linker was reacted with peptides bearing the complementary reactive aminooxy function to prepare the POC 10–12. The coupling reaction was carried out by simply mixing the two components without any additive, i.e. the oligonucleotide 9 and the desired peptide in slightly acidic conditions (pH 4.6). The POC 10 and 11 were obtained after HPLC purification in almost 50% isolated yields. The same process was further extended to the preparation of carbohydrate–oligonucleotide conjugate 12 [12]. Biologically relevant peptides and carbohydrate were used to prepare the conjugates. For instance, the arginine–glycine–aspartic acid (RGD) peptide is a cyclopentapeptide containing the RGD sequence which is known to be a powerful and selective ligand for αvβ3 integrin receptor. It has been studied for its tumour targeting property and ability to deliver DNA in the cells. Similarly, the NLS sequence is a nuclear localising signal peptide sequence derived from the simian virus 40 antigen. The carbohydrate–oligonucleotide conjugates can be useful to target the dendritic cells through their sugar binding receptors (lectins).
2.3 Oxime ligation for 3′ conjugation (Scheme 4)
Similar to the 5′ extremity, a post oxidation strategy was utilised for the generation of aldehyde moiety at the 3′ position. The commercially available modified solid support (1-dimethoxytrityl-3-fluorenylmethoxycarbonylaminopropane-2-succinoyl) long chain alkyl amino-CPG 13 was employed to incorporate the 1,2-amino-alcohol moiety in the oligonucleotides. This strategy was chosen because the oxidative cleavage of the 1,2-amino-alcohol moiety (such as serine residue) is a well-known strategy to generate the aldehyde function [13]. After the automated DNA synthesis, the oligonucleotides were cleaved from the support and deprotected by the conventional ammonia treatment to afford the oligonucleotide 14, bearing the 3′ amino-alcohol linker. The oligonucleotide 15, containing the 3′ aldehyde moiety was generated by the periodate oxidation. However, further studies with the solid support 13, showed that it gives irreproducible results and the efficiency of the support for the introduction of 1,2-amino alcohol was found to decrease over a period of time even when stored at low temperature. Consequently, a new commercially available solid support, 3-[(4,4-dimethoxytrityoxy)-glyceryl-1-succinoyl]-long chain alkyl amino-CPG 16, was preferred for the introduction of the 3′ aldehyde by the oxidative cleavage of the intermediate 1,2-diol moiety [7b]. The 3′ aldehyde containing oligonucleotide was used to prepare the oligonucleotide conjugates with RGD or NLS peptides, carbohydrate and fluorescent reporter groups similar to the one discussed for 5' conjugation [7].
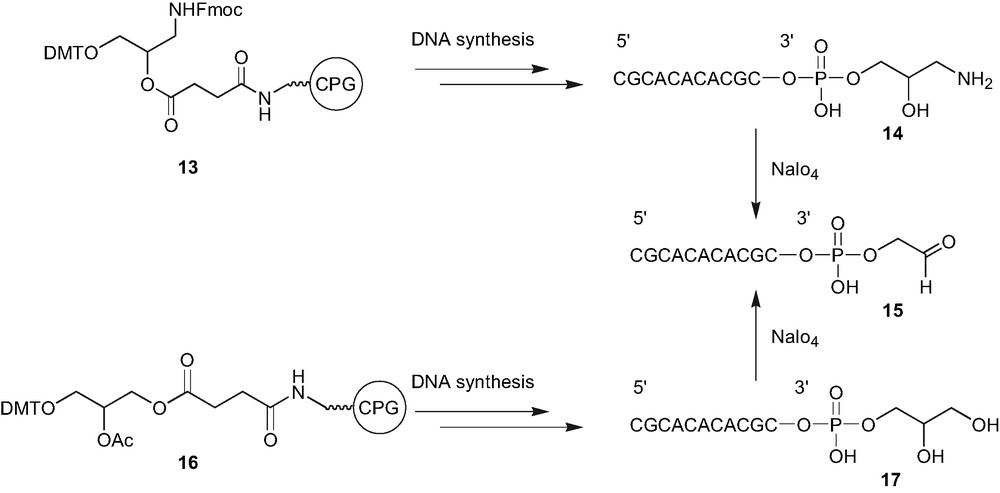
Synthesis of 3′ aldehyde containing oligonucleotide 15.
2.4 Oxime ligation for 2′ conjugation
A useful application of oxime ligation was recently reported by Zatsepin et al. [14]. They showed the possibility of using the 2′-OH position of the ribose moiety in the oligonucleotide to anchor multiple peptide sequences through the oxime ligation. Oligonucleotides carrying 2′ aldehyde groups were synthesised from a diol precursor. Coupling reaction with peptides containing a N-terminal aminooxy moiety afforded POC sustaining single or multiple peptides in good yield. Modification at the 2′ position is chosen due to the fact that 2′-O-alkyloligonucleotides improves the binding affinity for the target RNA [15].
2.5 3′,5′ Bifunctionalisation (Schemes 5 and 6)
In the recent past, we have focussed our attention towards the bifunctionalisation of oligonucleotides. The question before us was that if the aldehyde function can be generated at the two ends separately, whether it would be possible to do the same at both the terminus simultaneously. This would be important because it would give the ability to modify both the ends of oligonucleotides to counter the sensitivity towards the nucleases and also to induce various combinations of desirable properties by anchoring reporter molecules of choice. This could indeed be done as has been shown in Scheme 5 [16]. The oligonucleotide synthesis was carried out using the solid support 16, and the phosphoramidite linker 7, was introduced during the last step of automated DNA synthesis. Usual deprotection with ammonia followed by acetic acid treatment gave the oligonucleotide 17, carrying a diol function at both the extremities. The aldehyde was then generated by the periodate oxidation leading to 3′,5′ bis aldehyde oligonucleotides 18. This precursor was utilised to prepare the oligonucleotide conjugate 19 and 20, bearing either RGD or NLS peptides at both ends. Similarly, 3′, 5′ bis-carbohydrate–oligonucleotide conjugates were also prepared.
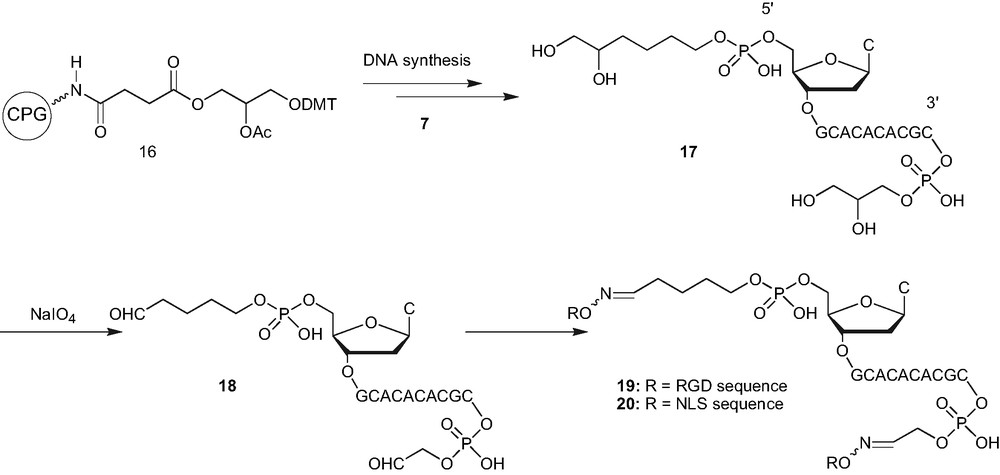
Preparation of 3′,5′ bis conjugates with similar groups.

Preparation of 3′,5′ bis conjugates with different groups.
However, the above mentioned strategy has only a limited application because it does not permit the preparation of bis-functionalised conjugates bearing two different groups. The preparation of such oligonucleotide conjugates would need a orthognal protection strategy so that the generation of aldehyde functionality and the subsequent conjugation at the two extremities could be carried out in a sequential fashion. We have recently reported a method to prepare bis-functionalised oligonucleotide conjugates bearing different groups at the two extremities based on sequential formation of the oxime bonds (Scheme 6). The said strategy is utilised to prepare conjugates bearing two different peptides or a peptide and a fluorescent reporter group [17].
Another important problem of interest to us was that whether it is possible to prepare oligonucleotide bearing the aminooxy function at one end and the aldehyde function at the other end of the same strand. This would be interesting because such oligonucleotide with sticky ends would then cyclise through the formation of intramolecular oxime bonds. Our recently published results show that this can certainly be done [18]. Scheme 7 shows the preparation of cyclic oligonucleotide derivatives 22a–h. The results are important because such oligonucleotides are known to possess a strong resistance to the exonucleases, as they do not have any end for the digestion process to begin. Besides, these have excellent DNA and RNA binding efficiency along with good strand displacement activity. Consequently, the development of synthetic strategy for cyclic oligonucleotides is a topic of current interest [19].

Preparation of cyclic oligonucleotides via intramolecular oxime bond formation.
2.6 Hybridisation properties
The hybridisation properties of conjugates for the target complementary sequence was investigated by melting temperature (Tm) measurements to evaluate the influence of the anchoring of the reporters groups on the stability of the duplex oligonucleotide. It was shown that incorporation of the reporters onto the nucleobase dramatically decreases the stability of the duplex. However, the modifications at the extremities do not perturb the binding affinity. Also, when modification is carried out by the peptide moieties containing the arginine or lysine residues, a moderate increase in the stability of the duplex is noticed.
3 Conclusion and perspectives
It can be therefore, concluded from the results and data available that the formation of oxime bonds can be utilised for efficient oligonucleotide conjugation, not only with peptides but a range of appropriately functionalised reporter groups including the carbohydrates, fluorophores or intercalants [20]. The main advantage of using the oxime ligation is the chemoselectivity. The investigations are also underway to further develop the method so as to prepare the cyclic oligonucleotide of longer length. We are now looking to explore the oxime ligation in new avenues such as for the anchoring of oligonucleotides on solid surfaces. This has potential application in the fascinating field of DNA microarray technology [9]. Furthermore, we are exploring the possibility of using the oxime ligation for the preparation of oligonucleotide conjugates bearing multivalent sugar residues (glycoclusters). Also, the possibility of using this ligation strategy for the preparation of templated oligonucleotide assemblies is being currently investigated.
Another aspect of the oxime ligation that needs attention is that though the oxime bonds are usually stable over a wide pH range, these can be hydrolysed under harsh conditions. This might not be desirable in many instances (aptamer strategy). Recently, we have developed and reported a new method to prepare the POC based on glyoxylic oxime bond formation [21]. The new method could be more suitable as the glyoxylic aldehyde is known to be more stable and so far not reported to react with the amino side chains in the peptides. It should be pointed out here that while the use of oxime ligation is attractive, there is a possibility of displacement reaction to occur [22]. However, we have not observed any such problem during the course of our investigations over past several years. Another problem with the use of oxime bonds could be that it exists in isomeric forms, E and Z. It is possible that the two isomers may show difference of properties. Nevertheless, the overall result makes us believe that more interesting results are yet to be explored. This also gives us hope that over a period of time, the oxime ligation would establish as a method of choice for the oligonucleotide conjugation.


