1 Introduction
There is growing interest in understanding and manipulating protein–protein interactions due to their central importance in many biological processes. Protein–protein interactions are known to play a critical role in the normal function of cellular/organelle structure, immune response, protein enzyme inhibitors, signal transduction, and apoptosis. Rational approaches towards the recognition of protein surfaces may provide better insights into how proteins exactly interact with one another and is an alternative to enzyme inhibitor design as a molecule-based disease therapy [1]. Functional group presentation on the surface of a protein is primarily divergent in nature. In binding protein surfaces, designed molecules must account for the extensive solvation of the protein surface, as well as its topology. Tight binding, therefore requires the involvement of large surface areas and multiple points of functionality. Numerous recent works show that these contact interfaces [2] (functional epitopes) are relatively small (8–12 residues), flat and that the binding affinity and the selective recognition highly depend on precisely located ‘hot spots’ [3], mostly solvent buried [4] and discontinuous, as well as on the plasticity of these interfaces. Despite important breakthrough, most notably in algorithm developments, reliable predictions of these interfaces remain elusive. New experimental approaches are necessary for the search of potential ligands. One of the present difficulties in the design of compounds targeting the surface interface is related to the nature, the size and the orientation of the substituents. Nowadays, no global approach allows the rational design of such molecules. However, combinatorial approaches have shown that small peptides can efficiently produce good complementary interface for the protein–protein contact surface [5,6]. Natural products such as cyclosporine, FK506 and rapamicine also display contact interfaces, which have been redesigned to alter their binding to cyclophilin or FK506-binding protein respectively [7,8]. We here propose a new approach that may easily provide potentially complementary surface with a given recognition interface. This approach (template-directed combinatorial surface) is founded on the concept of scaffold for tethering and directing chemical groups or molecules as recognition elements (hot spots), whose assembly provides modular molecular surface complementary to targeted binding interfaces (Scheme 1).
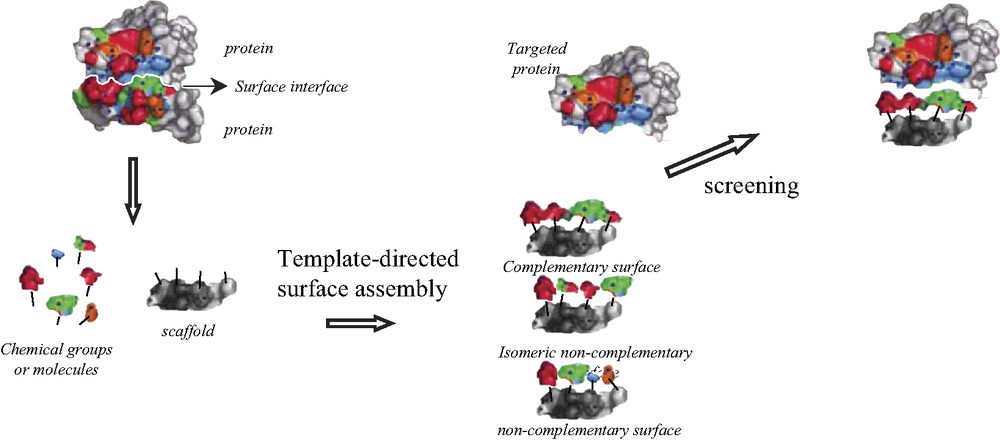
Template-directed combinatorial surface assembly principle.
This methodology allows to generate various types of surfaces as functional epitopes, in a combinatorial and/or sequential manner, from a subset library of functional groups and molecules that are independent from a structural point of view. It may provide a more global and more precise system to explore targeted surfaces by varying the nature of the scaffold and by adapting in an iterative manner the number and the diversity of the ligands assembled on the scaffold. For instance, from a given scaffold class and a given chemical strategy, the number of sites and the relative geometry can be varied (e.g. n elements on 4 attachement sites ⇨ n4 combinations) directly or by amplification following the dendrimer growing principle (e.g. n elements on 4 sites + 4 amplifications ⇨ n16 combinations).
This concept also allows the exploration of discontinuous surface complementarity, which is still difficult to address with classical approaches. In particular, tethering of functional epitopes to a scaffold may help to circumvent the problem of secondary and tertiary structures practiced by proteins to direct hot spots in the correct topology. A multi-entry combinatorial exploration (e.g., within the recognition elements and relative to their repartition on the scaffold) is allowed to adjust and refine the recognition. One can expect from this flexibility to be able to select a molecule capable of distinguishing its target between homologous proteins whose active sites only differ by few amino acids. Furthermore, our approach is evolutive because it will benefit from a directed chemical evolution of the hits through iterating of the selection process as well as the choice of the chemical nature of the recognition elements by analogy with phage display screening.
2 Results and discussion
At the crux of the approach is the linking of different components onto the scaffold, which has to be highly efficient as well as versatile (e.g. compatible with a large molecular functionalities). We selected the oxime bond as a chemoselective ligation tool between an oxyamine function and an aldehyde. This ligation technique is highly efficient, compatible with a wide variety of chemical functions and allows the oxime bond formation between unprotected fragment without any coupling reagent and with minimal chemical manipulation [9,10]. We selected Regioselectively Addressable Functionalized Template (RAFT) as a suitable scaffold to direct the combinatorial assembly of the components on one face and to present a reporter group on other face for detection purpose [11,12]. RAFT molecules represent topological cyclic decapeptides containing orthogonally protected attachment sites pointing to opposite faces of the template backbone that are chemically accessible at the gram scale. They display homogeneous conformation control that has been successfully exploited for protein mimicry [13,14] or surface functionalization [15,16]. In addition, we recently [11,17] reported the successful chemoselective oxime assembly of amino-oxy-carbohydrates or cyclopeptide to RAFT molecule for the presentation of recognition motifs clusters to direct independently and separately the ligands as well as the reporter groups [11,12].
2.1 Synthesis of the RAFT
Synthesis of the RAFT scaffold is performed on the G-Sasrin® resin using the Fmoc/tBu strategy [18] (Scheme 2). Glycine at C-terminal end was essential to secure the subsequent cyclization step with the N-terminal lysine α-amino group without epimerization.

Linear peptide synthesis of the RAFT.
After deprotection of the N-terminal Fmoc protecting group and mild acid cleavage of the resin, the peptide is cyclized [11] in solution in 82% yield. The Alloc protecting group is removed using the well-established Pd0/PhSiH3 procedure [19] and the corresponding free Lysine side-chain sulfonylated with dabsyl chloride in 76% yield. Then, the remaining Boc-protected Lysine side-chains were smoothly cleaved with 50% trifluoroacetic acid in dichloromethane at room temperature and the corresponding amino groups coupled with BocSer(OtBu)OH (Scheme 3) as glyoxylaldehyde precursors. Removal of the Boc/tBu groups was achieved quantitatively by treatment with TFA. Oxidative cleavage of the amino-alcohol moiety of seryl residue with sodium periodate [20] afforded the desired four Nɛ-glyoxylyl-lysyl containing cyclodecapeptide 1 in 51% overall yield after RP-HPLC purification.

Dabsyl-functionalization of the RAFT.
2.2 Design and synthesis of the ligands
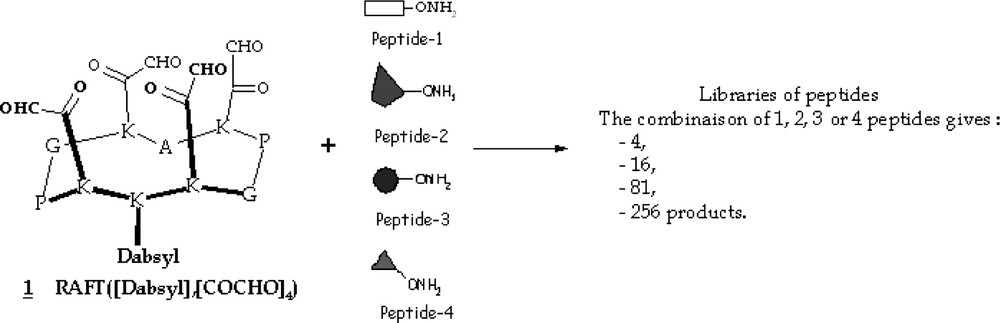
As a paradigm, we chose to target the avidin protein surface in order to validate the template-directed combinatorial surface assembly concept and to study the combinatorial ligations on the RAFT-scaffold. Several streptavidin or avidin binding peptides exhibiting high affinity were isolated using chemical as well as biological combinatorial approaches. From these peptide sequences, we designed four different linear and negative charged-bearing peptides, functionalized with an amino-oxy function for assembly [21].
The four peptide sequences were synthesized via a solide-phase strategy using a Fmoc/tBu chemistry. Those were designed to be of different lengths in order to have different masses and retention times affording an easier detection (HPLC or ES-MS). Moreover, all N-terminal peptide sequences were ended with a glycine residue to provide an identical reactivity during the oxime bond formation and thus avoid such a biais during the generation of the libraries (Scheme 4).

The four designed peptides.
2.3 Study of the combinatorial formation of oxime ligation on a scaffold
In our study, we assembled four different peptides to the four attachment sites of the RAFT scaffold through oxime bond formation. This provided theoretically 256 different molecules. This mixture could be differentiated in four subsets according to the following peptide combinations: one subset containing the four assemblies presenting only one peptide (e.g. RAFT([peptide-i]4)); one subset containing the 84 combinations of the RAFT presenting two different peptides (e.g. RAFT([peptide-i], [peptide-j])); one subset containing the 144 combinations of the RAFT presenting three different peptides (e.g. RAFT([peptide-i], [peptide-j], [peptide-k])) and one subset containing the 24 combinations of the RAFT presenting four different peptides (e.g. RAFT([peptide-i], [peptide-j], [peptide-k], [peptide-l])).
We synthesized the four-peptide-library (one experiment, 256 molecules) and all the possible sub-libraries were then obtained by combining the peptides by 1 (four experiments, one molecule by experiment), 2 (six experiments, 16 molecules by experiment) and by 3 (four experiments, 81 molecules by experiment) (Scheme 5).
The different libraries synthesized.
Scheme 6 presents an example of one of the simplest libraries containing one component (here peptide-1). This reaction allowed us to set up the best reaction conditions for the oxime coupling, e.g. 6–24 eq. of the amino-oxy peptide to consume all the amount of RAFT present in the reaction medium and make it easier the purification of the mixture after completion of the reaction. An ammonium acetate buffer was chosen and the reaction lasted on average 24 h at r.t.
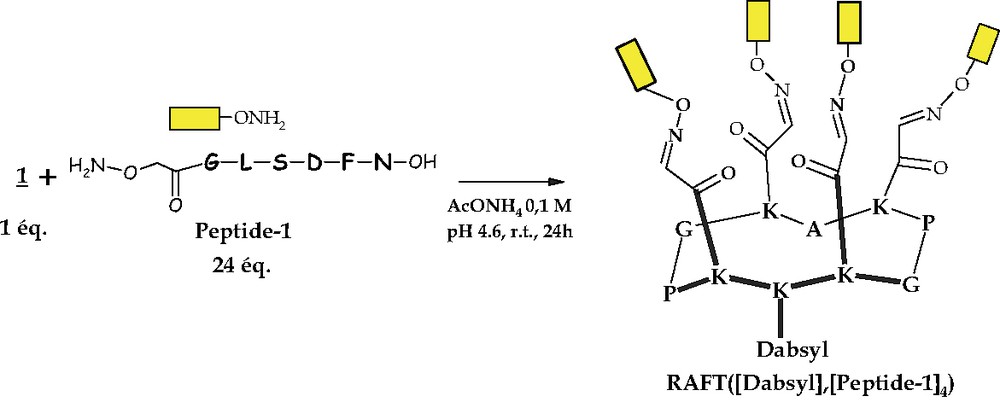
An example of the product of a reaction through chemoselective ligation.
In that way, it was possible to quickly synthesize all the 15 possible combinations (Scheme 5), which could be obtained by combining the four different peptides with 1.
2.4 Characterization of the coupling reaction
For a better clarity, we present here an illustrative case of library based on the assembly of two components, namely peptide-1 and peptide-3. It is anticipated to obtain 16 different molecules overall corresponding to all the possible compositions statistically possible by assembling the peptides on the RAFT. These molecules can be distributed into five subsets according to their compositions in peptides assembled onto the RAFT molecule:
- • two subsets, corresponding each to either peptide-1 or peptide-3 assembled alone on the RAFT, namely RAFT[(peptide-1)4] and RAFT[(peptide-3)4], providing for each subset one molecule;
- • two subsets, corresponding each to peptide-1 and peptide-3 assembled by 3 on the RAFT, namely RAFT[(peptide-1)1, (peptide-3)3] and RAFT[(peptide-1)3, (peptide-3)1], providing for each subset four isomeric molecules;
- • one subset, corresponding to peptide-1 and peptide-3 assembled by 2 on the RAFT, namely RAFT[(peptide-1)2, (peptide-3)2], providing six isomeric molecules.
Scheme 7 presents the characterization by HPLC and ES-MS of the libraries. As shown, five signals are obtained as anticipated. The corresponding integrations provide a ratio of 1:4:6:4:1 as expected as well. Moreover, the differences in retention times obtained by HPLC for the different groups are in agreement with the composition of the molecules (e.g. compounds containing the more polar peptide-1 are more polar compared to those containing peptide-3). From these observations one can conclude that the ratio follows the statistical distribution predicted above. This suggests that the chemical reactivities of the two peptides are the same towards the oxime bond formation and that the library obtained is not biased.
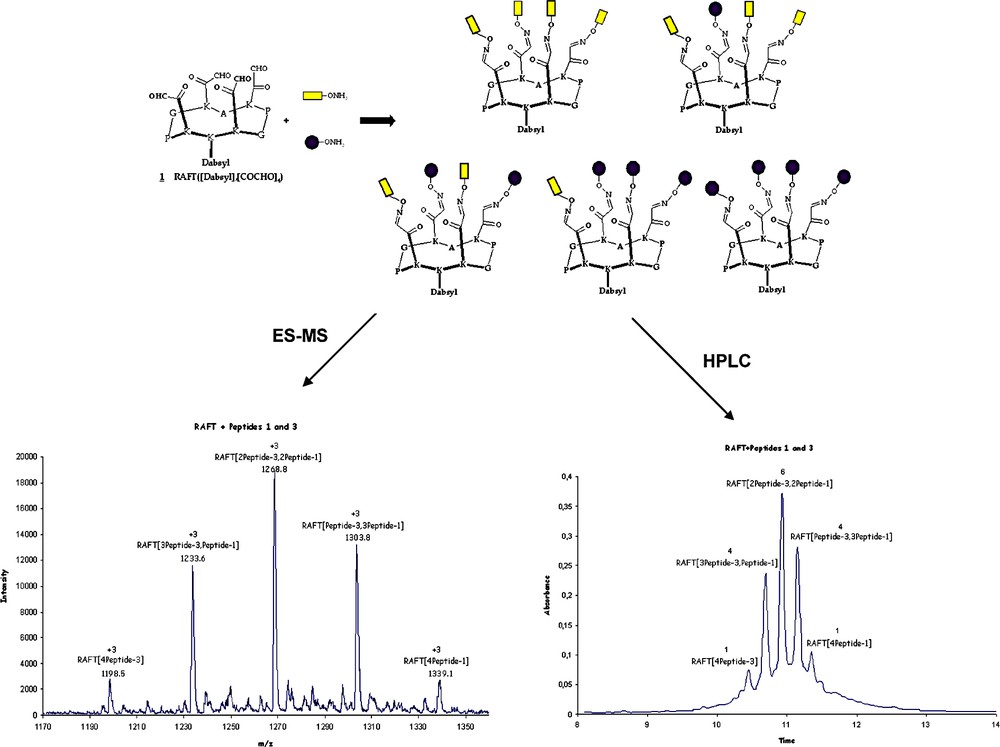
Example of the characterization of a reaction.
All the libraries composed of two different peptides obtained so far displayed the same behavior. However, it is important to remember that the resolution observed by HPLC relies mainly on the difference in peptide retention time which is not always that much different. For libraries based on the assembly of three and four components, the analysis was more complex but deconvolution could be possible, at least on the basis on the mixture observed with two components. Most of the components were characterized by ES-MS for all the libraries so far but for some of them, LC-MS coupling detection will be required. Work is currently under progress to this extend.
2.5 Test of the libraries
After having synthesized and characterized the different peptide libraries and sub-libraries, we performed tests on affinity chromatography [22]. In order to assess the potential recognition of the libraries to avidin, we used an avidin-agarose bead. The different peptide libraries presenting a dabsyl dye [23] on the lower face of the scaffold were poured on the column where the non-recognized molecules were not retained and the recognized ones were bound giving a red color to the bead. Matrix analysis of the different results obtained with the library and the sub-libraries with their composition should provide the composition in peptides assembled to RAFT required to display affinity towards the target. The preliminar results indicated that some mixtures exhibited a better affinity than others towards avidin or avidin-biotin complex. Work is in progress to characterize and analyze the corresponding molecules.
3 Conclusion
The chemical part of our project is validated since the reaction mixtures are easy to prepare, to purify and obtained within a day. The oxime bond ligation is well suited to realize combinatorial assembly of chemical groups derivatized by amino-oxy function on a given scaffold-aldehyde. It provides an easy method to obtain all the combinations of the chemical groups in a row as well as all the related sub-composition. The analysis of the libraries using HPLC and ES-MS techniques demonstrated that the libraries are not biased since the product distribution follows the statistical predictions. Finally the preliminary column affinity tests carried out in this study are encouraging since some libraries displayed recognition properties towards avidin, works are currently in progress to establish the composition of the ligands.
For the future, our aim will be to enlarge the types of molecules used to realize the different surface mimetics like new libraries based on constrained peptides (e.g., cyclopeptides) as well as on organics ligands (e.g., carbohydrates, heterocycles…) targeting proteins for drug discovery.


