1 Introduction
Solution of novel macromolecular structures is usually done by a variation of the heavy-atom method, utilizing the X-ray scattering properties of a small number of heavier atoms, whose scattering signal stands out from the contribution of the bulk of lighter carbon, nitrogen, and oxygen atoms building up the biopolymers. This signal may be provided by the large number of scattering electrons, as is the case in the single or multiple isomorphous replacement (SIR or MIR) methods, by the anomalous diffraction contribution of certain atoms in the single- or multi-wavelength anomalous diffraction (SAD or MAD) techniques, or by a combination of both (SIRAS, MIRAS: single or multiple isomorphous replacement with anomalous scattering) [1,2].
In the SIR or MIR approach, it is advantageous to use the heaviest atoms possible, and the most popular derivatization reagents contain elements from the last rows of the periodic table, such as Pt, Au, Hg, and U, often in the form of complex ions, such as, for example, PtCl62–, Au(CN)2–, HgI42–, and UO22+. Native protein crystals can be either soaked in solutions containing these compounds, or they can be present in the crystallization liquor. In the MIR method, the differences between reflection amplitudes measured from the derivatized and the native crystals are used first to locate the heavy atoms and then to estimate the protein phases, leading to the initial, hopefully interpretable, electron density map.
The MAD approach makes use of the anomalous scattering properties of selected atoms, which change abruptly if the wavelength of X-rays is varied in the vicinity of the X-ray absorption edge of these atoms. In this method, the native diffraction data are not necessary, since all data are measured from one crystal, usually at three wavelengths, corresponding to the peak, edge, and remote point of the X-ray absorption (or fluorescence) curve. The SAD method is based on the data measured at only one wavelength and relies exclusively on the Bijvoet differences between the intensities of the centrosymmetrically related reflections.
The contribution of the heavy atoms to the total diffraction signal can be estimated from the following formula [3]:
In this formula, ΔF is the isomorphous |FH – FN| difference; fH is the atomic scattering factor of the heavy atom; and NH and NP are the number of the heavy and all-protein atoms in the crystal structure, respectively. A similar formula applies to the anomalous signal [4], and the expected average Bijvoet ratio is:
One fully occupied mercury atom with 80 electrons and f" value of about 10 electron units near its absorption edge may provide the following signals:
in lysozyme with MW = 13.3 kDa <ΔFiso>/<F> = 53% <ΔFano>/<F> = 6.6%
in ribosome with MW = 2.3 MDa <ΔFiso>/<F> = 4% <ΔFano>/<F> = 0.5%
In practice, the diffraction intensities are measured with the accuracy not better than a few percent, and the weak heavy-atom signal can easily be lost in the noise. Clearly, it is advantageous to have several heavy-atom sites for phasing structures of larger proteins.
One of the ways to incorporate into protein crystals multiple heavy-atom sites is to utilize clusters consisting of several metal atoms bound together and providing a large number of scattering electrons, possibly with a significant anomalous diffraction signal.
Several proteins, mainly among those involved in the redox or electron transfer pathways, contain natural multi-atom clusters of such transition metals as Fe, Ni, Cu, Mn, Mo [5]. These intrinsic clusters may contain some inorganic ions, for example sulfides, cyanides, carbon monoxide or water, and are coordinated by sulfhydryl groups of cysteine (mainly Fe) or imidazole groups of histidine (mainly Cu). Classic examples of simple metalloproteines are ferreedoxins and high-potential iron proteins (HIPIPs), containing iron–sulfur clusters of various types [6]: digonal [Fe2S2], tetrahedral [Fe4S4] or pseudo-tetrahedral [Fe3S4]. Trinuclear copper centers exist in ceruloplasmins and laccases [7]. The nitrogenase complex contains three different types of metal clusters, including the unusual molybdenum–iron–sulfur coenzyme [8,9]. The intrinsic metals can be conveniently used for phasing crystal structures of such proteins, based on their anomalous scattering properties.
2 Diffraction properties of metal clusters
The use of metal clusters offers a simple way of introducing to protein crystals multiple heavy-atom sites, thereby enhancing the achievable phasing signal. However, the phasing effect of metal clusters does not simply correspond to the accumulation of the contributions of individual metal atoms. At low resolution, when the individual metal positions are not resolved, the whole cluster acts as a ‘superatom’, whose diffraction contribution corresponds to the sum of the total number of (anomalous) electrons within the cluster. Since the reflection intensities are proportional to the square of the number of the scattering electrons, the cluster consisting of N atoms of Z electrons each provides, at low resolution, a contribution proportional to (N Z)2, which is higher by N than N (Z)2, delivered by N individual scatterers.
Fig. 1 illustrates the contributions estimated for the octahedral Ta6 and the tetrahedral Fe4 clusters at various resolution ranges and Fig. 2 shows the anomalous difference Fourier synthesis calculated for the Ta6 cluster at high, medium and low resolution. At low resolution, when the clusters act as superatoms, they significantly enhance the scattering signal, up to the expected factor of (N)1/2 at the zero diffraction angle. However, at the intermediate resolution, in the range extending between one and two inter-metal distances, the diffraction contribution of the cluster is smaller than that of the sum of the individual scatterers. When the diffraction data resolution exceeds the inter-metal distance and the individual sites become resolved, their signal increases again [10]. The use of metal clusters, is therefore, beneficial at low as well as at high resolution, but is not very helpful at the intermediate resolution range.
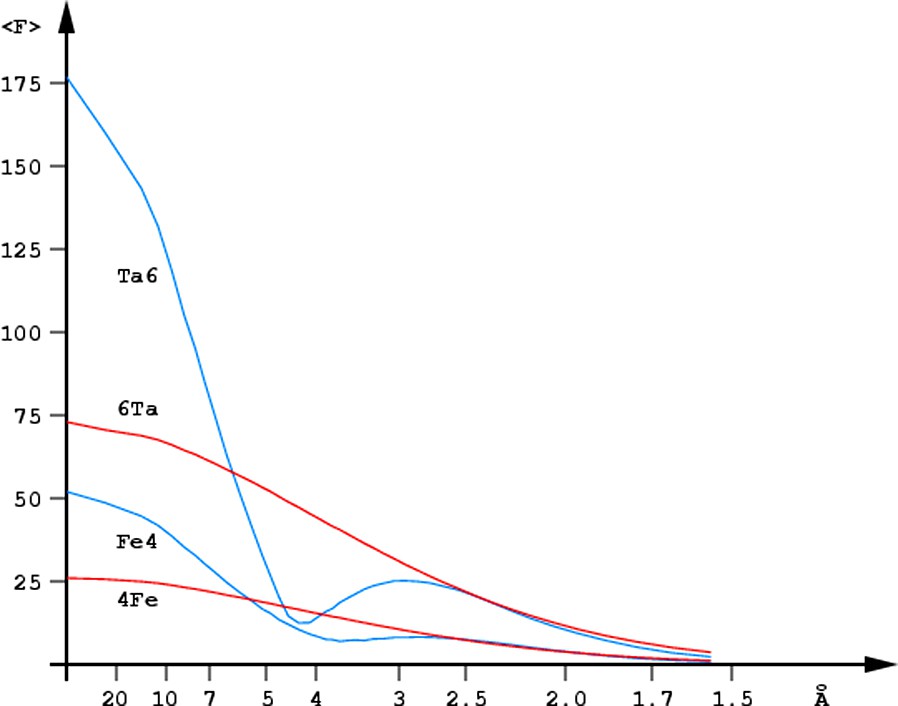
Resolution dependence of the scattering contribution of the Ta6Br12 cluster (Ta6, blue) and of the six randomly positioned Ta atoms (6Ta, red) as well as the analogous dependence for the Fe4S4 cluster (Fe4, blue) and four isolated Fe atoms (4Fe, red).
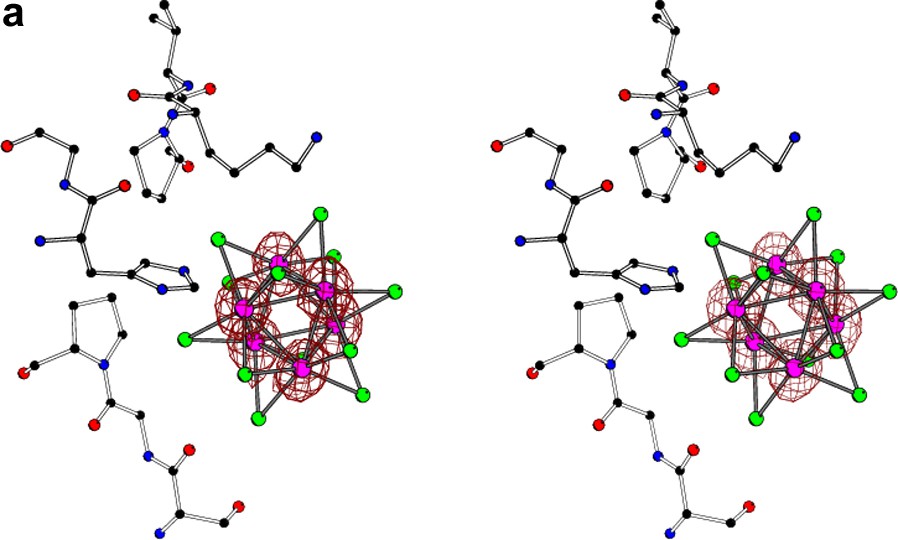
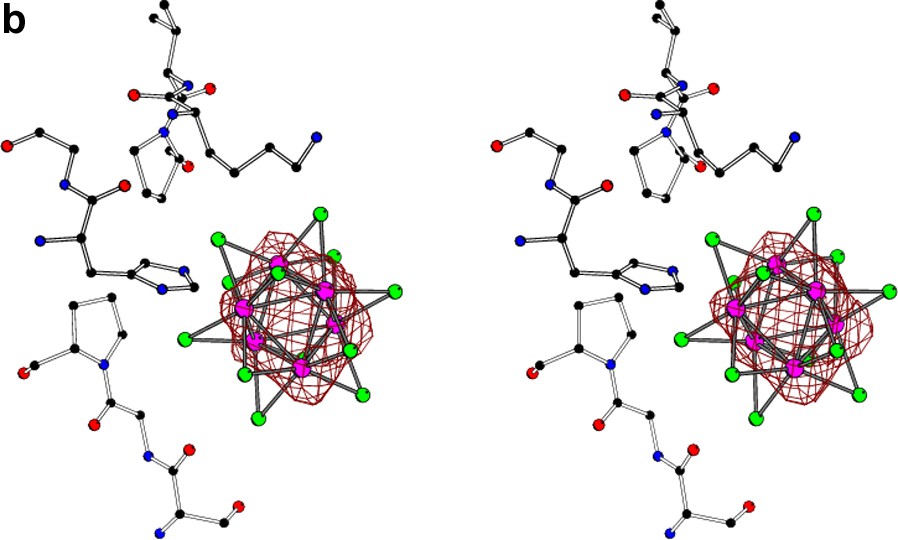
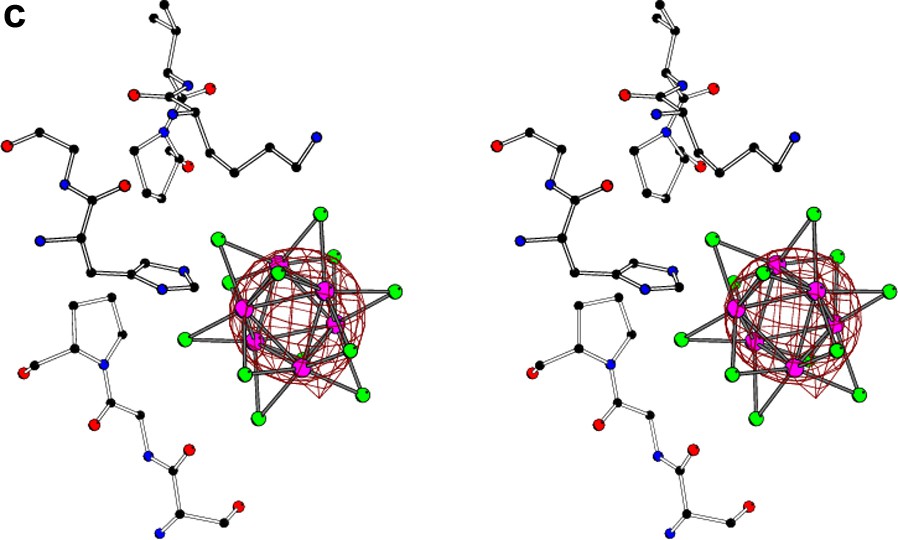
The stereo view of the Ta6Br12 cluster located near the surface of the molecule of glucose isomerase [10] with superimposed anomalous difference Fourier map (at 1σ level) calculated at (a) 1.3-Å, (b) 3.5-Å, and (c) 5.0-Å resolution. At high resolution, the individual atoms of tantalum are resolved, at 3.5 Å the orientation of the cluster can still be deduced, but at 5.0 Å resolution the whole group of six tantalum atoms behaves as a spherical superatom.
3 Chemical requirements
To be successfully used for phasing macromolecular crystal structures, metal clusters should fulfill certain criteria. They have to be stable in aqueous solutions that, beyond the protein itself, often contain various inorganic and organic additives used as stabilizers, buffering agents, or precipitants. Crystallization liquors may contain simple salts, most frequently ammonium sulfate, sodium or lithium chloride, potassium phosphate, or sodium formate. The organic precipitants are often polyols or their oligomers, such as polyethylene glycol, 2-methyl-2,4-pentanediol, various sugars, or glycerol. Buffers used for maintaining the pH value range from simple salts based on, for example, phosphates or acetates, to such organic compounds as TRIS (tris-(hydroxymethyl)-aminomethane), HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), ADA (N-(2-acetamido)iminodiacetic acid). Some proteins are crystallized in the presence of small amounts of such additives as dioxane, isopropanol, or (especially membrane proteins) amphiphilic detergents. The variation of crystallization conditions is enormous, since different proteins require a specific content of the mother liquor for a successful crystal growth and their stable survival. The reagents used for heavy-atom derivatization should not actively interfere chemically with other compounds present in the crystallization liquid.
Several cluster compounds, to a large extent fulfilling the above criteria, have been applied for phasing protein crystal structures, Table 1. The idea of using a metal cluster for this purpose was first suggested in the early 1960s by Linus Pauling. He established the three-dimensional structures of the octahedral clusters of niobium and tantalum Nb6Cl122+ Ta6Cl122+ [11] and proposed that they could be used to produce the isomorphous derivative crystals of the hen egg-white lysozyme. However, this attempt was not successful [12], and the crystal structure of lysozyme was solved a little later [13,14] in the laboratory of David Phillips by using simpler heavy-atom reagents.
Examples of cluster compounds used in macromolecular crystallography
| Symbol | Formula | Ref |
| TAMM | C(HgOCOCH3)4 | [52] |
| PIP | [(en)PtI2Pt(en)]2– en = NH2CH2CH2NH2 | [18] |
| TaBr | Ta6Br122+ | [43–45] |
| Au11 | [Au11(CN)3(PAr3)6(PAr′3)] | [53,54] |
| Ir4 | Ir4(CO)8(PA3)3(PA2A′) | [34] |
| W11Rh | [PW11O39{Rh2(CH3COO)2}]5– | [35] |
| W18As | [(AsW9O33)2(SnPh)4] | [35] |
| W12 | [PW12O40]3– | [55,56] |
| W11 | [PW11O39]7– | [38] |
| W12Si | [SiW12O42]8– | [38] |
| W17 | [P2W17O61]10– | [38] |
| W18 | [P2W18O62]6– | [57,58] |
| W30 | [NaP5W30O110]14– | [59] |
Since they provide systems of very high electron density, heavy metal clusters are routinely used as contrasting agents in cryo-electron microscopy [15].
4 Examples of the use of clusters for phasing
Because of their high phasing power at low resolution, the polynuclear metal clusters often have been used for solving crystal structures of large proteins or their multicomponent complexes. Fig. 3 illustrates the three most popular clusters used for that purpose.
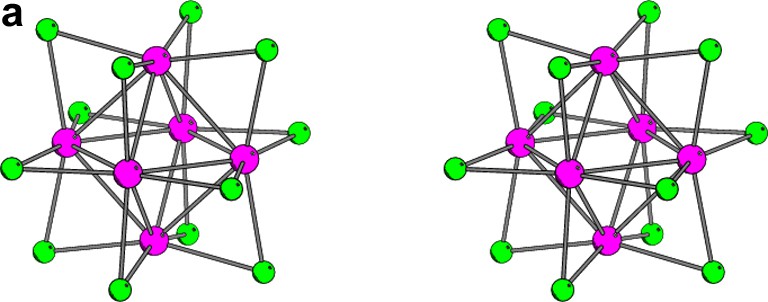

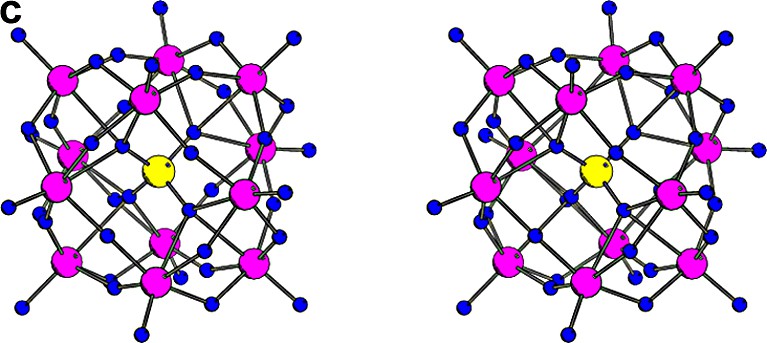
The stereo illustrations of the most popular clusters used for phasing protein crystal structures. (a) Ta6Br12 cluster of Oh symmetry, where the diameter of the regular octahedron of tantalum atoms is about 4.1 Å. (b) Au11 cluster of C3v symmetry, where the green ligands represent the cyano (or halide) groups and the yellow ligands represent the tri-arylphosphine groups. The diameter of the gold cage is about 5.3 Å. (c) W12 cluster of Td symmetry with the diameter of the skewed cuboctahedron of the tungsten atoms is about 7.3 Å.
The nucleosome core particle with 206 kDa in the asymmetric unit was solved [16] using two cluster compounds, TAMM (tetrakis(acetoxymercuri)methane [17]), and PIP (di-μ-iodobis(ethylenediamine)-di-platinum nitrate [18]). TAMM was used in solving the photosynthetic reaction center [19], the idiotype–anti-idiotype complex [20], and the glutathione S-transferase [21].
TAMM and PIP do not have direct metal–metal bonds, and therefore, are not strictly polynuclear cluster compounds, but their properties and practical use are similar to the proper polynuclear metal clusters.
A number of metal clusters played important roles in solving the structures of various ribosome particles. The Yonath group [22,23] tried many clusters in their work on the 50S particles of 1.5 MDa molecular weight [24–28], as well as for phasing the small 30S subunit of 850 kDa [29,30]. Some of them had specially designed ligand functions (maleimide or aminoethyl groups) for covalent binding to cysteine sulfhydryls in proteins (e.g. Au11 or Ir4 clusters [31–34]).
The Steitz group utilized the TaBr, W11Rh, and W18As clusters to successfully phase the structure of the large 50S ribosomal subunit at low resolution [35,36], as well as single-metal reagents at higher resolution of 2.4 Å [37].
The small 30S ribosomal subunit of 850 kDa was phased in the Ramakrishnan laboratory with the use of the TaBr, W12Si, and W17 clusters [38] at low resolution, and the phases were extended to 3 Å with the help of the single-metal compounds [39,40].
In the structure elucidation of the complete 70S ribosome particles with a molecular weight of about 2.5 MDa by the Cate group [41], soaks in many cluster compounds tended to degrade the diffraction properties of the crystals, but the TaBr cluster proved useful in extending the initial cryo-EM phases, which were further extended by the use of the single-metal osmium and iridium derivatives [42].
The TaBr cluster has been used to solve many other protein crystal structures, and its usefulness has been chiefly promoted in the laboratories of Robert Huber in Munich [43,44] and of Gunther Schneider in Uppsala and Stockholm [45]. Table 2 gives examples of the structures solved with TaBr, which is the most widely used cluster compound and which can be synthesized in many different ways [46–51].
Structures solved using the Ta6Br122+ cluster as one of the derivatives
| Structure | PDB | MW in ASU | Resol. of | Ref. |
| Code | (kDa) | phases (Å) | ||
| Transketolase | 1NGS | 150 | 5.5/2.5 | [60] |
| Proteasome | 1PMA | 700 | 7.0/3.4 | [61] |
| GTP cyclohydrolase I | 1GTP | 510 | 6.0/3.0 | [62] |
| Dimethyl sulfoxide reductase | 1DMS | 90 | 2.2/1.9 | [63] |
| l-Fucose isomerase | 1FUI | 390 | 6.0/2.5 | [64] |
| Yeast RNA polymerase II | 1IQ3 | 514 | 4.5/3.1 | [65,66] |
| Conjugative coupling protein | 1E9R | 50 | 4.5/2.4 | [67] |
| Tricorn protease | 1K32 | 720 | 4.2/2.0 | [68] |
| γ-Complex of polymerase III | 1JR3 | 200 | 3.5/2.7 | [69] |
| l-Rhamnulose-1-phosphate aldolase | 1GT7 | 600 | 6.0/2.7 | [70] |
| Chromosome maintenance complex | 1LTL | 190 | 5.5/3.1 | [71] |
| Furin | 1P9 J | 440 | 4.0/2.6 | [72] |
| Photosystem II from T. vulcanus | 1IZL | 640 | 5.5/3.7 | [73] |
| Photosystem II from T. elongatus | 1S5L | 640 | 3.8/3.5 | [74] |
5 Conclusions
Solving novel crystal structures of macromolecules always involves the use of heavy atoms that serve as markers for the initial phasing through their isomorphous or anomalous scattering signal. Their signal has to stand out from the background of the scattering of all other lighter atoms (C, N, O, H) present in the biological material. For large systems it may be not enough to use simple heavy-atom derivatization reagents; however, polynuclear metal clusters are ideally suited for this purpose, since they provide a strongly enhanced phasing signal, particularly at low resolution. The current advances in molecular biology make it possible to isolate, purify, and crystallize large biological complexes, having a molecular weight of several megadaltons, and the usage of various cluster compounds in macromolecular crystallography is expected to increase in the future.


