1 Introduction
Purple photosynthetic bacteria have evolved an elegant solution to the problem of harvesting solar energy to power their photosynthesis. Their solution is based upon a rather simple modular construction system (see Refs. [1–3] for recent reviews). Heterodimeric units, consisting of αβ-apoprotein pairs which non-covalently bind two or three molecules of bacteriochlorophyll a (Bchla) and one or two molecules carotenoid (Car), aggregate to produce circular holocomplexes. The apoproteins serve as a protein ‘scaffold’ which precisely organises the light-absorbing pigments for optimal energy transfer.
Most purple bacteria contain two basic types of light-harvesting complexes, called LH1 and LH2. These complexes can be readily released from the photosynthetic membrane by suitable detergent treatment, and then independently isolated and purified [1]. As a result of this they have been intensively studied and we now know a great deal about their structure and function [4].
2 The structure of LH2 from Rhodopseudomonas acidophila strain 10050
Fig. 1 shows the absorption spectrum of the LH2 complex from Rhodopseudomonas acidophila. Monomeric Bchla in organic solvents, such as 1:1 v/v acetone and methanol, absorbs in the NIR at ~770 nm. This Qy band shifts to the red when Bchla is incorporated into the antenna complexes and the position of this absorption band depends upon the pigment's environment [5]. The two major LH2 NIR absorption bands, at 800 and 850 nm, reflect the presence of two populations of Bchla within the holocomplex. The ‘three fingered’ absorption bands between 450 and 550 nm are due to the Car. In this case they are all rhodopin-glucoside, which has 11 conjugated double bonds. The LH2 complex from Rps. acidophila contains two types of small, hydrophobic apoproteins. The α-apoprotein contains 53 amino acids and the β-apoprotein contains 49. In 1995 the first X-ray crystal structure of this LH2 complex was described, to a resolution of 2.5 Å [6]. This has now been improved, by cryocooling the crystals, to a resolution of 2 Å with a final R-factor of 17.3% (RCSB PDB ID code 1NKZ) [7].
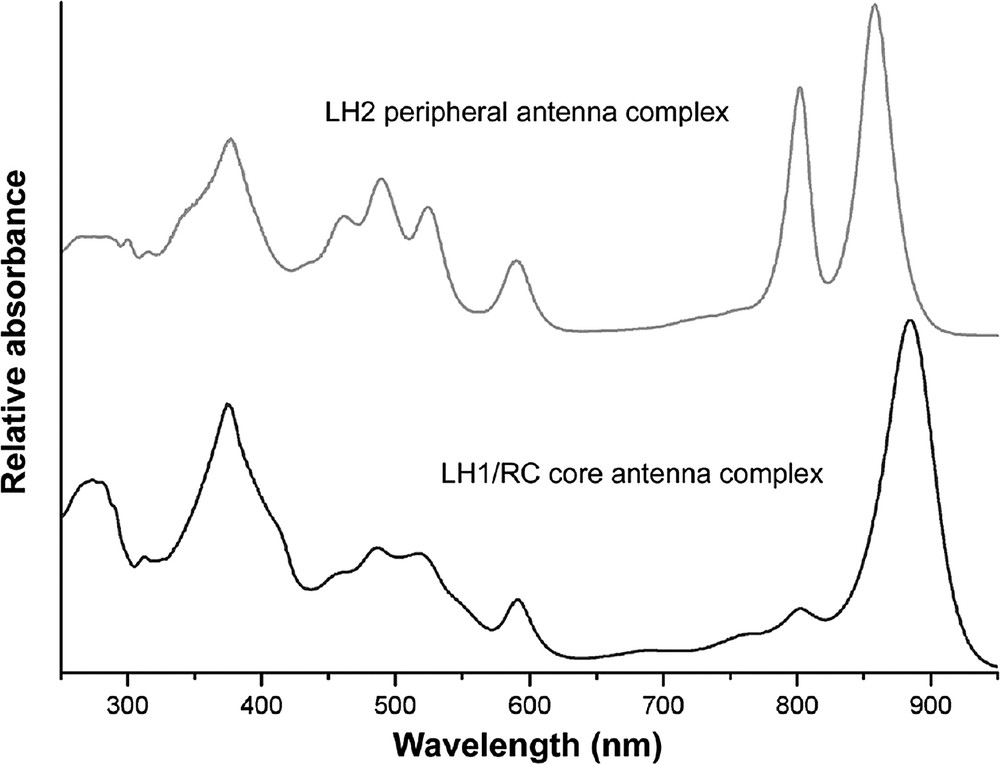
The absorption spectrum of the LH2 and the RC-LH1 ‘core’ complexes isolated from Rps. acidophila.
The LH2 complex is a circular nonamer (α9β9) (Fig. 2). It is a cylinder where the inner walls are made up of nine α-apoprotein α-helices and the outer walls are composed of nine β-apoprotein α-helices. All the pigments (with the possible exception of a second Car molecule per αβ pair) are located between these two ‘rings’ of transmembrane α-helices. The N-and C-terminal of the apoproteins fold over at the top and bottom of the cylinder, interacting with each other to firmly enclose the pigments.
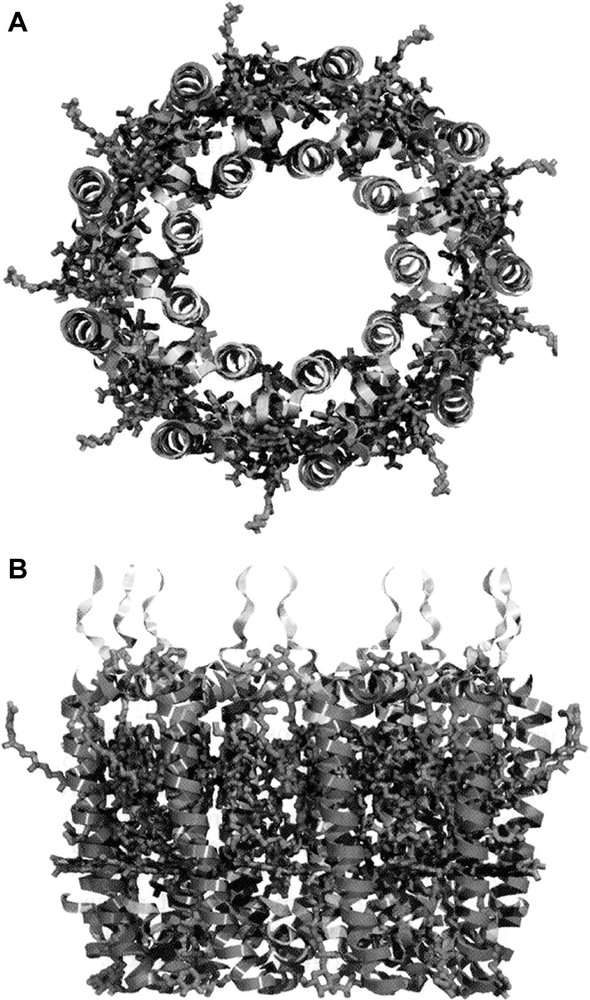
The overall structure of the LH2 complex from Rps. acidophila.
A – A ‘top-view’.
B – A ‘side-view’.
The pigments (shown as wire frames) are sandwiched between the helices (ribbons) of the apoproteins. The second Car molecule can be easily distinguished as it sticks out from the rest of the structure. For a colour representation of this Figure the reader is advised to consult Fig. 2 of Ref. [2].
Two groups of Bchla's (predicted from the absorption spectrum) are clearly seen in the structure (Fig. 3). On the N-terminal side of this complex (the cytoplasmic side). Here is a ring of nine monomeric Bchla molecules, one per αβ pair. The bacteriochlorin rings of these Bchla molecules lie flat, parallel to the plane of the photosynthetic membrane, and are separated centre to centre by 21.2 Å. Their central Mg2+ atoms are liganded to a carboxylate extension of the N-terminal methionine residue of the α-apoprotein. These Bchla molecules absorb at 800 nm and are called the B800 Bchla’s.

The organisation of the Bchla molecules in the LH2 complex from Rps. acidophila.
Top – The B800 Bchla molecules.
Bottom – The B850 Bchla molecules.
Both views are taken looking down on the LH2 structure from the N-terminal side of the complex.
Towards the C-terminal end of the transmembrane α-helices – there is a second ring of 18 tightly-coupled Bchla molecules. These absorb at ~ 850 nm and are called the B850 molecules. The B850 bacteriochlorin rings lie parallel to the transmembrane α-helices, that is perpendicular to the membrane plane. Their central Mg2+ atoms are liganded to conserved histidine residues (one coming from the α-apoprotein and one from the β-apoprotein). This means that each αβ pair binds two of these Bchla molecules. The B850 bacteriochlorins are strongly exciton coupled and their Mg2+–Mg2+ distances are 9.5 Å (within each αβ pair) and 8.8 Å (between neighbouring αβ pairs). The closest approach of the B850 bacteriochlorins rings to each other is 3.9 Å (within each αβ pair) and 3.6 (between neighbouring αβ pairs).
The single well-ordered Car per αβ-pair runs through the whole depth of the complex. It starts on the N-terminal side of the complex in one αβ pair and then passes in van der Waals contact past the edge of the B800 bacteriochlorin ring. It then proceeds to crossover into the next αβ-pair and passes (again in van der Waals contact) over the face of the bacteriochlorin ring of the B850 Bchla, which is liganded to the α-apoprotein. This Car is in a twisted all trans configuration and, because it spans and interacts with 3 separate α–β pairs, it acts to ‘bolt’ the structure together. Indeed it most cases the absence of this Car results in the LH2 complex not being stably assembled [8]. The improved 2.0 Å structure of LH2 has tentatively suggested the presence of a second Car molecule per αβ-pair, all be it in rather low occupancy. In this case the Car is located on the outside of the complex. The sugar head group is at the periplasmic side (opposite to the orientation of the well-ordered Car) and about half of the conjugated chain is well ordered. The middle of the molecule has two cis bonds that have been cautiously located at C12 and C15 in the current model; thus the end sticks out away from the complex. It is not clear whether this represents the ‘native’ conformation or whether it has been partially ‘pulled away’ by the detergent used to solubilise the complex from the photosynthetic membrane. Further work is needed to really establish the status of this putative second Car molecule per αβ-pair.
A year after the crystal structure of the LH2 complex from Rps. acidophila was first described the structure of the LH2 complex from Rhodospirillum molischianum was also determined [9]. This complex is highly homologous to the LH2 complex from Rps. acidophila. The main difference is that the Rhodospirillum molischianum complex is an octamer. It is still not clear what controls the oligomeric size of these antenna complexes or whether there is a functional consequence of the different ‘ring’ sizes. Readers that are interested in a detailed comparison of the arrangement of the pigments in these two types of LH2 complex are advised to see Ref. [10] and in particular Fig. 8 in that paper.
3 The structure of the RC-LH1 ‘core’ complex
Fig. 1 shows the absorption spectrum of a typical RC-LH1 core complex. Unlike the LH2 complexes the RC-LH1 core complex only has a single large Bchla absorption band in the NIR. The LH1 complex is constructed on the same modular principle as the LH2 complexes [1–3]. It is an oligomer of αβ-apoprotein pairs, where each pair only binds two Bchla molecules (called B875) and one or two Car molecules. Both these Bchla molecules have then Mg2+ atoms liganded to the conserved histidine residues, just as in the case of the B850 Bchla molecules in the LH2 complexes. Early EM pictures of intact photosynthetic membranes from Rhodopseudomonas viridis had clearly shown that the LH1 complex surrounds the RC [11].
Until quite recently the highest resolution picture of an LH1 complex had come from an 8.5 Å projection map of a reconstituted LH1 complex from Rhodospirillum rubrum [12]. This map was generated by electron crystallography on 2D crystals of the LH1 complex. It showed a closed ‘ring' with 16 αβ pairs. There was extra density between the modelled apoproteins which was proposed to represent the B875 Bchla molecules. This suggested that if each αβ-pair bound two Bchla molecules then LH1 contains 32 Bchla’s. Karrasch et al. pointed out that the hole in the middle of the LH1 ring was large enough to accommodate the RC. This model has now received strong support from a number of EM studies, for example see [13].
However, as described in detail in [1], there has been and continues to be much debate on the number of αβ-pairs in the LH1 ring. Why should this be such an issue? The answer comes from an apparent problem due to the properties of the electron transfer reaction within the RC. Following two turnovers the RC produces a fully reduced ubiquinol molecule which has to transfer its reducing equivalents to the cytochrome b/c1 complex, as part of cyclic electron transport. If the RC is surrounded by a palisade of rigid α-helices from the LH1 complex how can the ubiquinol ‘escape’ to find the cytochrome? This issue has been extensively reviewed in [1].
Two species of purple bacteria, Rhodobacter sphaeroides and Rhodobacter capsulatus, contain a protein called Puf X [14,15], which appears to be associated with the LH1 complex and is able to facilitate ubiquinol/ubiquinone exchange between the RC and the cytochrome b/c1 complex.
Very recently we published the crystal structure of the RC-LH1 complex from Rhodopseudomonas palustris at a resolution of 4.8 Å [16] (Fig. 4). Even though the resolution of this structure is limited it does reveal a lot of interesting information. This RC is surrounded by an oval, rather than a circular, LH1 complex consisting of 15 αβ pair and their associated Bchla molecules. The elliptical LH1 complex has approximate outer dimensions of 110 by 95 Å. The orientation of the long axis of the ellipse coincides with the long axis of the RC and allows the LH1 complex to wrap tightly around the RC. The LH1 complex is prevented from completely encircling the RC by a single transmembrane helix (called W). W replaces an αβ-pair and is located out of register with the other 15 αβ pairs. Still very little is known about W but it has been speculated that it is equivalent to the Puf X protein seen in the two Rhodobacter species. W is located directly in front of the Qβ binding site in the RC and is therefore ideally placed to provide a point of escape for the ubiquinol. This structure is tantalising. It suggests the answers to the Q problem but higher resolution data are urgently needed to fully confirm or refute this model.
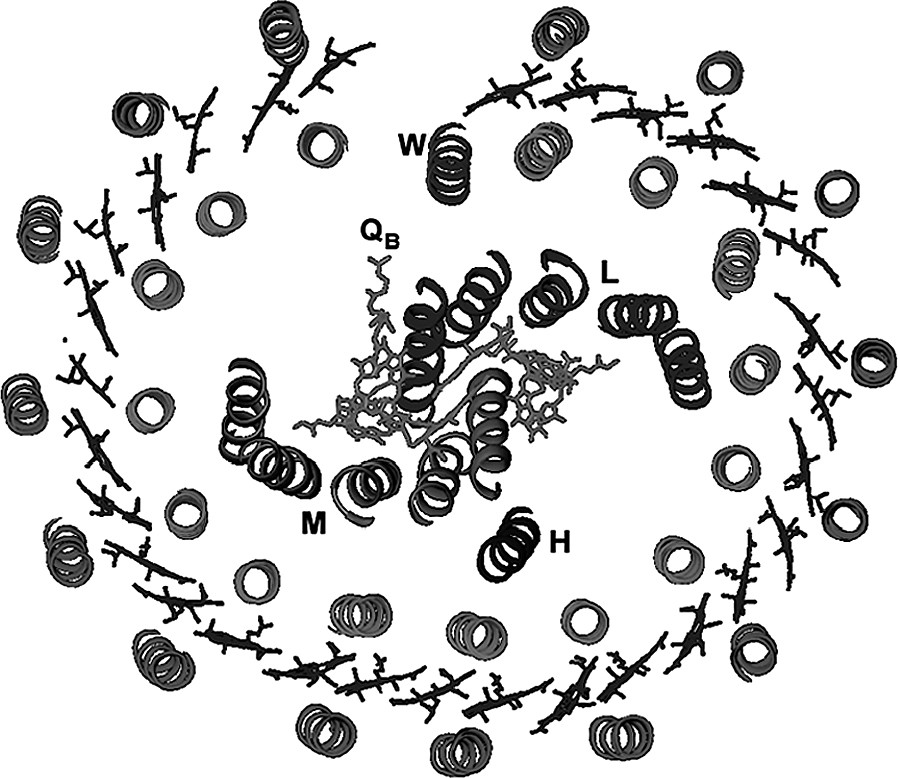
A plan of the structure of the RC-lh1 ‘core’ complex from Rps. palustris. The LH1 oval surrounds the RC. The three RC subunits L, M and H and the Qβ are labelled. Protein ‘W’ interrupts the regular LH1 oval opposite the Qβ binding site in the RC.
AFM studies on intact membranes for Rhodopseudomonas viridis, which contain regular RC-LH1 complex arrays, have clearly shown that in vivo the RC-LH1 complex is oval and not circular [17].
4 Energy transfer in the purple bacteria photosynthetic unit
In contrast to chlorophyll-containing antenna complexes from plants and algae, the purple bacterial antenna complexes are particularly suited to investigations of their energy-transfer properties since the different group of pigments, seen in their structures, are well separated in the absorption spectrum. This clear spectral separation means that they can each be independently excited by light so that the energy transfer from one group to another can be clearly distinguished. As a consequence of this all of the possible energy transfer reactions have been time-resolved from a few femto seconds to longer times [4,18].
A full discussion of these energy transfer reactions is beyond the scope of this short review. Fig. 5, however, summarises the currently available data for the major Bchla to Bchla energy transfer steps that take place from the periphery of the photosynthetic unit into the RC. Anyone wishing to read about the energy transfer reactions involving the carotenoids should consult the recent excellent review by Polivka and Sundström [19].
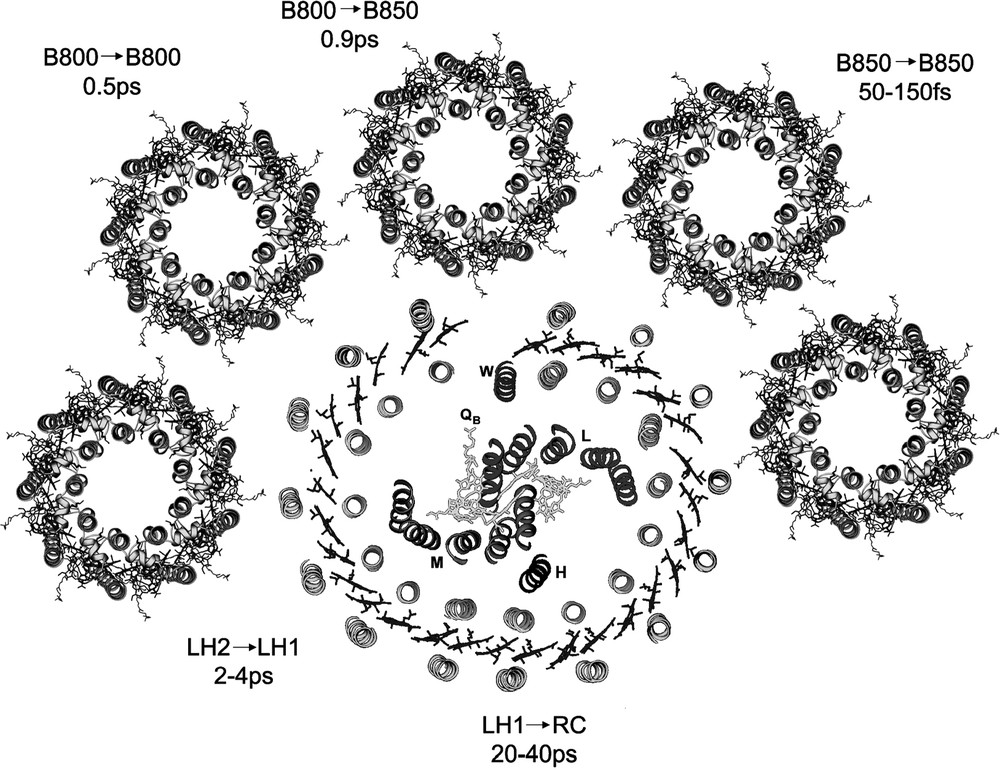
A cartoon of the purple bacterial photosynthetic unit with the times of the major Bchla → Bchla energy transfer reactions indicated. This is a hypothetical model of the PSU designed to help the reader visualised the various ET steps that have been time-resolved. It is not meant to represent how these complexes are organised relative to each other in the natural photosynthetic membrane.
Acknowledgments
This work was supported by the BBSRC, the Wellcome Trust and NEDO.



Vous devez vous connecter pour continuer.
S'authentifier