1 Introduction
Oxygenic photosynthesis supplies all higher life on earth with food and produces all of the oxygen in the atmosphere by a process that converts the light energy from the sun into chemical energy. Three different kinds of organisms are able to perform oxygen-evolving photosynthesis: plants, algae and cyanobacteria. The primary steps of photosynthesis have been conserved over 1.5 billion years of evolution remaining essentially the same in plants, algae and cyanobacteria.
The initial steps of photosynthesis, the light reactions, occur on a membrane system known as the thylakoids. Two large membrane protein complexes, photosystems I and II (abbreviated as PS I and PS II, respectively) are embedded in this membrane. They catalyze the initial step of photosynthesis, the light induced charge separation across the photosynthetic membrane. Both photosystems are functionally coupled by a pool of plastoquinones, the cytochrome b6f complex (which is another large membrane protein complex), and plastocyanin (cytochrome c6n replaces plastocyanin in same cyanobacteria). The photosynthetic membrane (hereafter referred to as the thylakoid membrane) is charged by this process (comparable to the charging of a battery), during which a proton gradient develops across the membrane. The electrical gradient and the proton gradient (i.e. the electrochemical gradient) drive the synthesis of ATP. Reduced hydrogen for sugar formation is provided by reduction of NADP+ and H+ to NADPH. The electrons for the entire photosynthetic electron transport chain are provided by photosystem II through the process of water oxidation.
2 Photosystem I
2.1 Overview of the function of photosystem I
Photosystem I is a giant membrane protein complex: the cyanobacterial PS I consists of 12 protein subunits to which 127 cofactors (chlorophylls, carotenoids, FeS clusters and phylloquinones) are non-covalently bound. Photosystem I catalyzes the light-driven electron transfer from the soluble Cu-containing protein plastocyanin on the lumenal side (i.e. the inside of the thylakoids) to ferredoxin at the stromal side (outside) of the thylakoid membrane. There exists some flexibility in terms of the soluble electron donors and acceptors. The heme containing soluble protein cytochrome c6 can serve as an alternate or unique electron donor to photosystem I in cyanobacteria. In plants, algae and cyanobacteria growing in iron-limited conditions, flavodoxin can act as the electron acceptor instead of ferredoxin.
Photosystem I from the thermophilic cyanobacterium Synechococcus elongatus is a trimer with a molecular weight of 1 056 000 Da. It is the largest and most complex membrane protein for which the structure has been determined [1,2]. It was crystallized by an approach that includes microgravity experiments, the determination of phase diagrams and seeding techniques [3]. Fig. 1 shows the image of the trimeric complex, revealing the complex organization of this nano-biosolar system. Each monomeric unit contains 12 proteins, 96 chlorophylls (the pigments that give plants their green color), 22 carotenoids (orange pigments), three 4Fe4S clusters, two phylloquinone molecules and four lipids.

The trimeric structure of photosystem I from cyanobacteria; the view direction is from the stromal side onto the membrane plane. The 12 proteins are shown in a backbone representation, with transmembrane helices shown as columns. The chlorophylls are represented by their chlorin head groups; their phytyl-tails have been omitted for clarity; the carotenoids are depicted in black.
The first step of the entire photosynthetic process is light capturing, performed by the large antenna system, which consists of 90 antenna chlorophylls and 22 carotenoids. The energy is transferred to the center of the complex, where the electron transport chain is located. When the energy excites a special pair of two chlorophylls P700, located in the center of the complex, charge separation occurs. Due to its absorption maximum at 700 nm, this pair of chlorophylls was named P700. The electron is transferred from P700* across the membrane by a chain of electron carriers. The electron is transferred stepwise from P700 to A (a chlorophyll a molecule), A0 (also a chlorophyll a molecule), A1 (a phylloquinone molecule) and from there to the three FeS clusters FX, FA and FB. From the terminal FeS cluster, FB, the electron is transferred to ferredoxin, which subsequently leaves the docking site to bring the electron to the Ferredoxin-NADP+-reductase, which then finally reduces NADP+ to NADPH. After this process has occurred once, P700+ has to be re-reduced to complete the reaction cycle. There is a docking site for soluble electron carrier proteins plastocyanin/cytochrome c6, which is located at the lumenal site of the complex, just underneath P700 (see Fig. 2B).

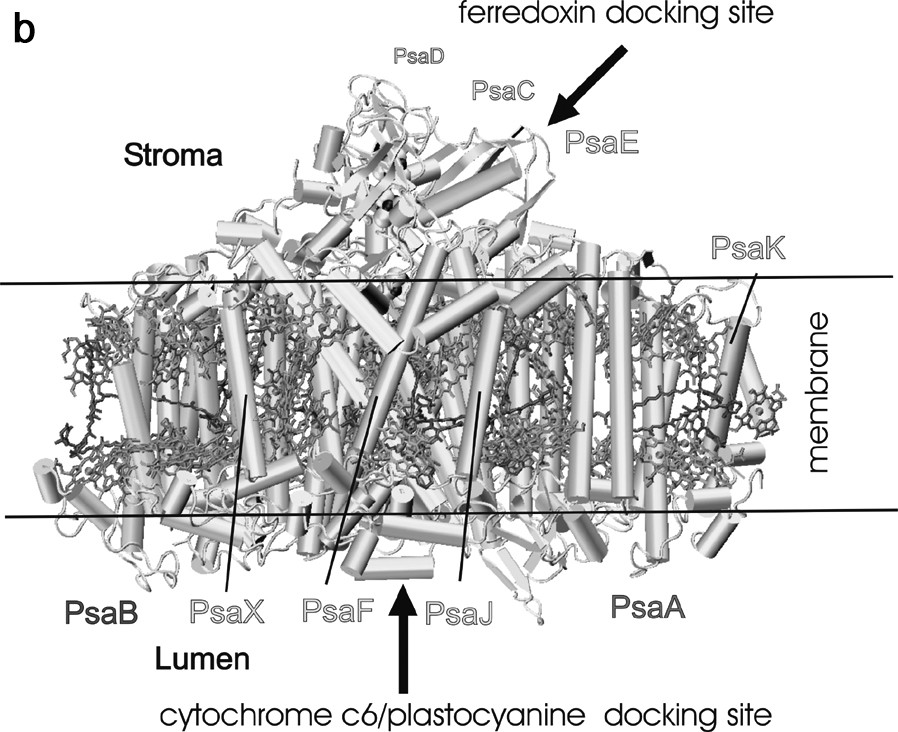
Arrangement of protein subunits and cofactors in one monomeric unit of photosystem I. (a) view from the stromal side onto the membrane plane. The three stromal subunits and all loops have been omitted for clarity. Transmembrane helices of the membrane intrinsic subunits are represented as cylinders. (b) Side view of the monomer of the PS I complex. The view is parallel to the membrane plane. All cofactors have been omitted except the three FeS clusters. The three stromal subunits PsaE, PsaC, and PsaD form a stromal hump on top of the transmembrane region and provide the docking site for ferredoxin. The docking site for plastocyanin/cytochrome c6 is located at the lumenal indention at the bottom of the transmembrane region. The chlorophylls and carotenoids are represented as in Fig. 1.
2.2 Structure of photosystem I
2.2.1 The protein composition of photosystem I
Cyanobacterial photosystem I consists of 12 different protein subunits, with names ranging from PsaA to PsaX, according to their genes. Fig. 2 shows two different views of the protein and cofactor arrangement in one monomeric unit of photosystem I. The large subunits PsaA and PsaB are the two most important proteins; they harbor most of the antenna system as well as most of the cofactors of the electron transport chain from P700 to the first FeS cluster FX. The five C-terminal helices are located in the center of the PS I and surround the electron transfer chain, whereas the six N-terminal helices flank the inner core on both sites. They are arranged as a trimer of dimers and coordinate the majority of the antenna pigments in PS I. The six smaller membrane-embedded proteins (PsaF, PsaI, PsaJ, PsaK, PsaL and PsaX) surround the core formed by PsaA and PsaB. Their main function is the stabilization of the antenna complex. PsaL also serves an important structural role, as it is involved in the formation of the trimer. A further function of the smaller subunits may be as a means through which photosystem I interacts with external antenna complexes. Three extrinsic subunits, which do not contain any transmembrane helices, are located at the stromal site of the photosystem I complex (shown in Fig. 2B). These three subunits (PsaC, PsaD and PsaE) form a ‘stromal hump’ that extends the membrane by 35 Å and provides the docking site for the soluble electron carriers ferredoxin and flavodoxin. The central subunit PsaC carries the terminal electron acceptors, the two FeS clusters FA and FB.
2.2.2 The electro-transfer chain
The electron transport chain is located in the center of one monomeric unit of photosystem I. The structural organization of the cofactors of the electron transport chain is depicted in Fig. 3. The organic cofactors of the electron transfer chain are arranged in two branches. They are named the A and B branches, because most but not all cofactors of the A branch and B branch are coordinated by PsaA and PsaB, respectively.
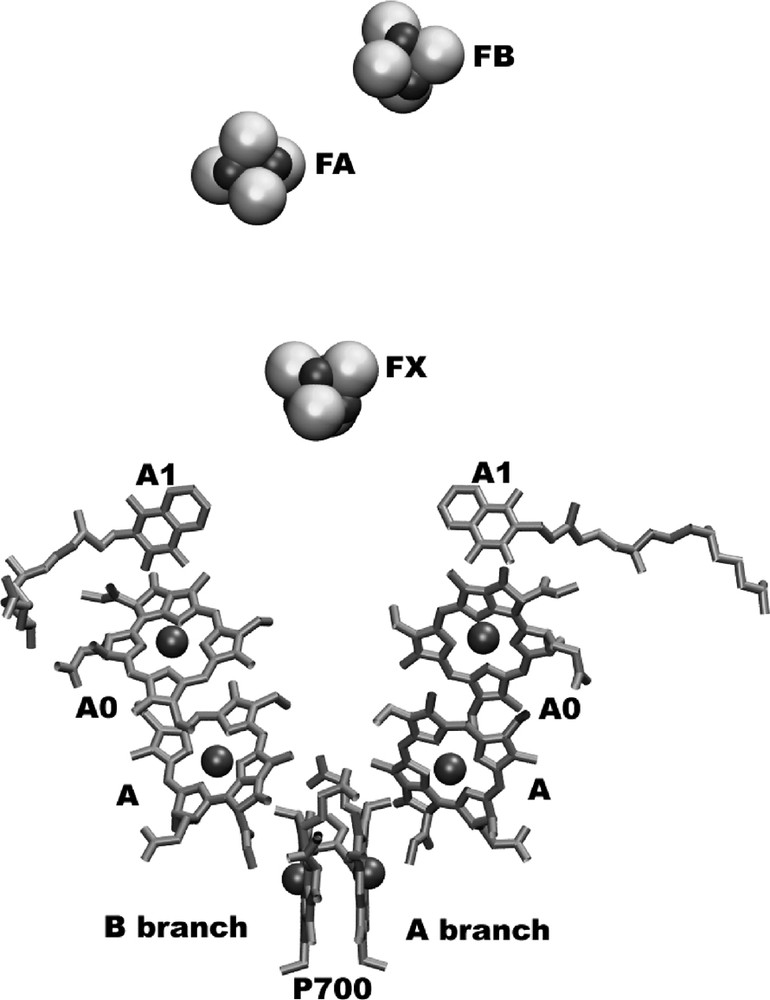
The electron-transfer system of PSI.
The view is parallel to the membrane plane. The organic cofactors of the electron transfer chain are arranged in two branches. The left branch is called the A-branch, whereas the right branch is the B-branch. The three FeS-clusters FX, FA and FB are located at the stromal side of the membrane (on top of the figure). The names of the cofactors are given, as derived from spectroscopic investigations.
2.2.3 The primary electron donor ‘P700’
The pair of chlorophylls assigned to the primary electron donor P700 is located close to the lumenal surface of photosystem I. P700 is comprised of two stereochemically different a-type chlorophyll molecules. The chlorophyll on the B-branch is a chlorophyll a molecule, stereochemically identical to all the other 95 chlorophylls in photosystem I, whereas the chlorophyll at the A-branch is chlorophyll a′, the epimer at the C13 position of the chlorin ring system. The protein environment further enhances the asymmetry of the two chlorophylls: three hydrogen bonds are formed between the protein and the P700 chlorophyll a' at the A-branch, whereas the P700 chlorophyll a at the B-branch does not form any hydrogen bonds to the protein environment [1]. The asymmetry could lead to significant differences in the redox potential of both chlorophylls, which may explain why the unpaired electron of P700+ is located to more than 85% on the B-branch chlorophyll [4]. A possible function of the asymmetry may be a gating of electrons along the two branches but to date this suggestion has not been proven experimentally.
2.2.4 The intermediate electron acceptor, ‘A’
From P700 the electron is transferred via one of the chlorophylls from the second pair of chlorophylls (A) to the first stable electron acceptor, A0. These first steps of electron transport are very fast (3 ps), such that the second pair of chlorophylls has not yet been detected by spectroscopy. The chlorophylls (‘A’) that may represent the primary electron acceptor are not directly coordinated by a ligand from the protein side chains, but by a water molecule, which forms the second H-bond to two asparagine side chains of the opposite branch. Recent work even suggests that they are not only involved in electron transfer, but that one of them may even represent P700* [5], but this topic is still matter of controversy.
2.2.5 The first stable electron acceptor, ‘A0’
The two chlorophylls located in the middle of the membrane may represent the spectroscopically identified electron acceptor A0. Their coordination is very unusual. In both cases, the sulfur atom of methionine provides the fifth ligand of the Mg2+, even if, due to the concept of strong acids and weak bases, there should be only a very weak interaction between sulfur in the methionine and Mg2+. It can be suggested that the weak interaction hinders a comparably stronger ligand (such as water) from entering the site, thereby leading to a chlorophyll which may behave as if it lacks a fifth ligand. This weak ligandation may be responsible for A0's extreme negative redox potential of –1000 mV and may be important for electron transport, given that the replacement of the methionine by histidine, which provides a strong ligand to the chlorophyll, blocks electron transport at the level of A0 [6].
2.2.6 The phylloquinones
The electron is further transferred from A0 to one of the phylloquinones, which represent the electron acceptor ‘A1’. The two phylloquinones are located at the stromal side of the membrane. The binding pockets are identical on both sites but differ significantly from all other quinone binding pockets found in proteins so far. Both quinones are π-stacked with a tryptophan residue and both show asymmetric hydrogen bonding: only one of the two oxygen atoms forms an H-bond to an NH backbone group, whereas the other oxygen atom is not hydrogen bonded at all. This could lead to a protein-induced asymmetry in the distribution of the unpaired electron in the radical state A1–•, formed when one of the phylloquinones is reduced. This may answer the question of why A1 has the most negative redox potential (–770 mV) of all quinones found in nature.
EPR studies and model calculations of vitamin K1–• (phylloquinone) in polar and unpolar solvents in comparison to PS I suggested that a very hydrophobic environment can cause a dramatic shift of the redox potential to the negative values. These are indicated by very large g-principal values in the EPR signals. The g-tensors for vitamin K1–• in an ether solvent shell are the same as that of A1–• in PS I [7]. These studies suggest a major role of a hydrophobic environment in causing a very negative midpoint potential in quinone radicals.
2.2.7 The FeS cluster FX
The electron proceeds from A1 to the FeS cluster FX. This FeS cluster is a rare example of an inter-protein FeS cluster, coordinated by four cysteines; two of them provided by PsaA and the other two cysteine ligands provided by PsaB. The loops of PsaA/PsaB that coordinate the FeS cluster FX are the best conserved sequence regions in PS I. In addition to its functional role in electron transport, FX is important in the structure and assembly of PS I, as PsaC, PsaD and PsaE can not be assembled into the PS I complex in the absence of FX. The FeS cluster FX needs several proteins (such as rubredoxin) for its assembly [8].
2.2.8 The terminal FeS clusters FA and FB
The terminal FeS clusters–FA and FB–are coordinated by the membrane extrinsic subunit PsaC. This subunit is located in the center of the stromal hump that provides the docking site for the soluble electron carrier protein ferredoxin. The cluster FA is more proximal to FX, whereas FB represents the terminal FeS cluster that transfers the electron to ferredoxin [9–11]. The redox potential of FB is less negative than the redox potential of FA, which leads to the fact that > 80% of electrons are located on FA rather than FB after one round of excitation and charge separation. The ‘trapping’ of the electron at FA, which is deeply buried in the protein matrix may be functionally important, as it may reduce the possibility of energy wasting and potentially dangerous side reactions (e.g., the reduction of other substances such as oxygen) by PS I. Recently, cocrystals have been grown that contain an active 1:1 super complex of PS I with ferredoxin [12]. EPR spectroscopic investigations on these crystals indicated a change of the electronic properties of FB upon docking of ferredoxin, thereby facilitating electron flow.
2.2.9 Are both electron-transfer branches active?
One of the most controversy-laden questions is whether one or both branches are active.
There is now experimental evidence indicating that this step can proceed on both branches, but at different rates. In the green alga Chlamydomonas reinhardtii, the electron transfer is approximately 50 times slower on the A-branch than it is on the B branch [13,14]. This could be the result of a higher activation energy barrier on the A- compared to the B-branch. This finding raises the question: why the difference? There is not a significant difference between the protein environment in both branches, but there are two lipid molecules located close to the pathway from A1 to FX that could be responsible for the asymmetry [1]. A negatively charged phospholipid against which the electron has to be transferred is located on the slower A-branch, whereas on the faster B-branch a neutral galactolipid has replaced the phospholipid. This could be one of the reasons for the higher activation energy barrier on the A- compared to the B-branch.
2.2.10 The antenna system
The antenna system of photosystem I contains 90 chlorophyll a molecules and 22 carotenes. The arrangement of the chlorophylls and carotenoids is shown in Fig. 4. The chlorophylls are the main antenna pigments. They capture light and transfer the excitation energy to the center of the complex, where the electron transfer chain is located. This energy transfer is highly efficient. After excitation of any of the antenna chlorophylls the chance that the energy is successfully transferred to P700 and that subsequent charge separation occurs is 99.98% at room temperature.
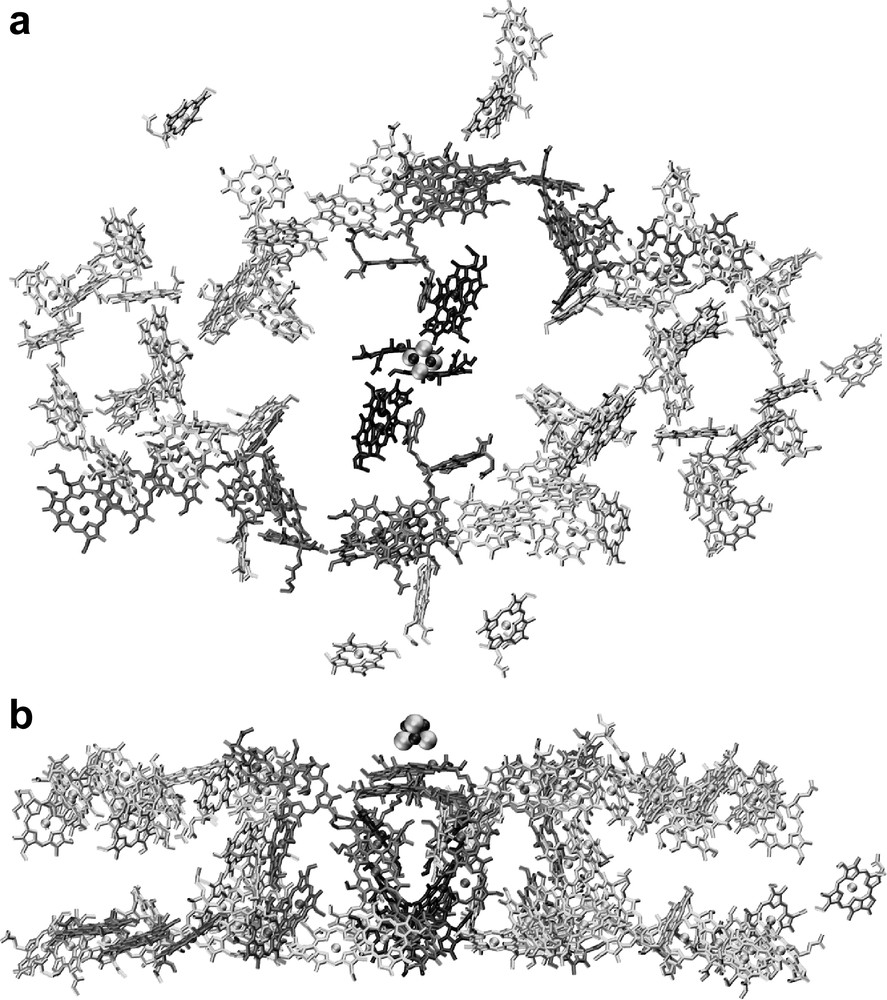
The antenna system of photosystem I. (a) Organization of the chlorophylls. The view is from the stromal side onto the membrane plane. For clarity, only the chlorin ring system of the chlorophylls is shown. (b) Organization of the chlorophylls; side view of the cofactors of PS I, the view is parallel to the membrane plane.
2.2.11 The chlorophylls
The arrangement of the antenna system in PS I is unique, as the chlorophylls form a clustered network. Each of the chlorophylls has several neighbors at a distance of less than 15 Å, so energy can be efficiently transferred to the center of the complex via multiple pathways. The system can be somewhat compared to the system of nerves in the brain in which a network of neurons is responsible for the high efficiency of information transfer. The antenna system in photosystem I is highly optimized for efficiency and robustness, as revealed by computational modeling studies based on its X-ray crystallographic structure [15,16].
The antenna system can be divided into a central domain, which surrounds the electron transfer chain, and two peripheral domains, flanking the core on both sides. In the peripheral domains, the antenna chlorophylls are arranged in two layers, one close to the stromal surface of the membrane and the other close to the lumenal surface of the membrane.
When a peripheral antenna chlorophyll is excited, the energy will first be transferred from this ‘two-dimensional’ layer to the central domain. In the central domain, the excitation energy can easily be exchanged between the two layers. From the chlorophylls of the central domain the excitation energy is then transferred to the electron-transfer chain. There are two chlorophylls that seem to structurally link the antenna system to the electron transfer chain. Recent mutagenesis studies on the ligands of these connecting chlorophylls showed alterations in the trapping time [17], but the question as to whether they play a crucial role in energy transfer is still not fully resolved.
2.2.12 The carotenoids
The carotenoids fulfill a triple function in photosystem I. First, they prevent the system from damage by over-excitation caused by excess light. In addition, they perform a structural role and function as additional antenna pigments [18]. Their photo-protective role is very critical not only for photosystem I, but for the entire photosynthetic electron transport. Chlorophylls are in principle dangerous and reactive molecules. Over-excitation can lead to the formation of chlorophyll triplets (3Chl), which can react with oxygen to form the toxic singlet oxygen, a very dangerous cell poison. The 3Chl triplet states are successfully quenched by transfer from 3Chl to 3Car, the latter one dissipating the excess energy by heat, thereby preventing photodamage. The system works very efficiently, even under high light conditions, as the 3Chl triplet state cannot be detected in the intact photosystem I. Multiple interactions can be observed between the carotenoids and the chlorophylls of the antenna system, which effectively protect PS I from photodamage.
2.3 Interaction of the antenna pigments within the PS I trimer
The trimeric nature of cyanobacterial PS I raises the question as to the functional importance of the trimerization. Trimers are preferentially formed under low light, and it has been shown that mutants lacking the subunit PsaL (which forms the trimerization domain) are growth-impaired under light limiting growth conditions, indicating that the trimeric organization may be critical for the PS I antenna system. There are several chlorophylls located at the interface between the monomers, which may be able to facilitate excitation energy transfer between the antenna systems within the trimer. Recent computational studies of the excitation energy transfer within the trimer indicate that the excitation energy transfer between monomers may occur much more frequently than expected, with the chance of trapping of the excitation energy transfer by a P700 from an adjacent monomer being 30% [19].
3 Photosystem II
3.1 Overview on the function of photosystem II
Photosystem II catalyzes the light driven electron transfer from water to plastoquinone. It provides the whole photosynthetic process with electrons by extracting them from water:
Thereby, oxygen is evolved and provided for respiration, which is the energy source for all higher non-photosynthetic organisms, including humans. With respect to photosynthesis, oxygen is only a by-product and, due to its high oxidation potential, is even dangerous for cells.
As in photosystem I, the light induced charge separation is initiated by the capturing of the light by an internal antenna system, which is much smaller in PS II than in PS I because it only contains approximately 40 antenna chlorophylls. The excitation energy is transferred into the center of the complex where the electron transfer chain is located. The excitation of a chlorophyll (or chlorophylls) with an absorption maximum at 680 nm leads to a charge separation across the photosynthetic membrane, where the electron is transferred to a mobile quinone, QB, via a chain of electron carriers that include a pheophytin and the tightly bound quinone QA. After two subsequent electron-transfer steps the doubly reduced QB–– binds two protons and leaves the binding pocket as PQH2. The empty pocket is then refilled by a plastoquinone from the PQ pool.
In each round of the photocycle, P680+ is re-reduced by extracting one electron from the Mn cluster, with a redox active tyrosine functioning as the intermediate electron carrier between P680+ and the Mn cluster. After four subsequent electron transfer steps, i.e. when four positive charges are accumulated at the Mn cluster, one molecule of oxygen is released.
Due to the high redox potential of P680+ of 1.1 V, photosystem II experiences severe photodamage. Side reactions, such as the direct oxidation of the protein by P680+ or by the formation of the 3P680 triplet and, subsequently, highly reactive singulet oxygen, lead to the irreversible damage of the core protein of PS II, D1, that binds most cofactors of the electron transfer chain including the Mn cluster. This protein has to be replaced every half an hour in bright sunlight. The repair cycle of PS II is a very exciting and intensively studied area of PS II research [20].
3.2 Structure of photosystem II
Photosystem II is a dimer in the photosynthetic membrane of both plants and cyanobacteria. It consists of 17 protein subunits to which 35–40 chlorophylls and 8–12 carotenoids are non-covalently bound. The first crystals of photosystem II that were able to split water have been grown from the thermophilic cyanobacterium T. elongatus [21] leading to the first X-ray structural model of the intact PS II complex at 3.8-Å resolution [22]. The crystals were competent in oxygen evolution [23] and their protein content has been analyzed in detail [22] (Fig. 5).
Comparison of the structural models of photosystem II at 3.8-Å [22] and 3.5-Å [26] resolutions. The pictures are sligthly modified after [22,26].
This model provided the first insight into the structure of the water-splitting photosystem II, and is the basis for the discussion of functional aspects with regard to the mechanism of water oxidation, electron transfer and the process of light capturing (Fig. 6).
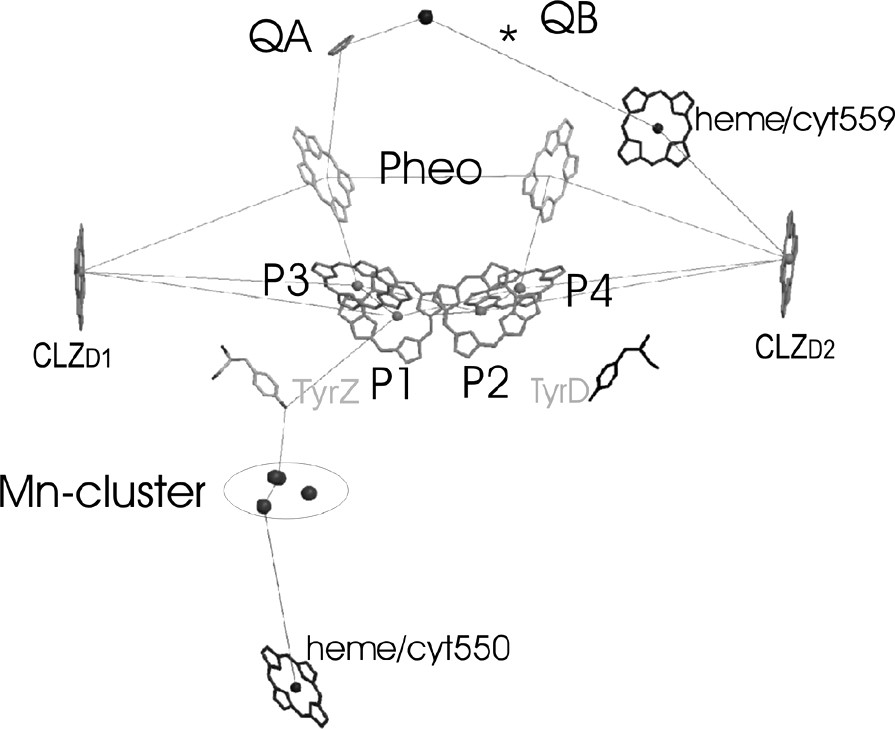
The electron-transfer chain of PS II in a side view, as revealed from the 3.6-Å structure of PS II from S. elongatus [33,24].
3.2.1 The protein subunits in PS II
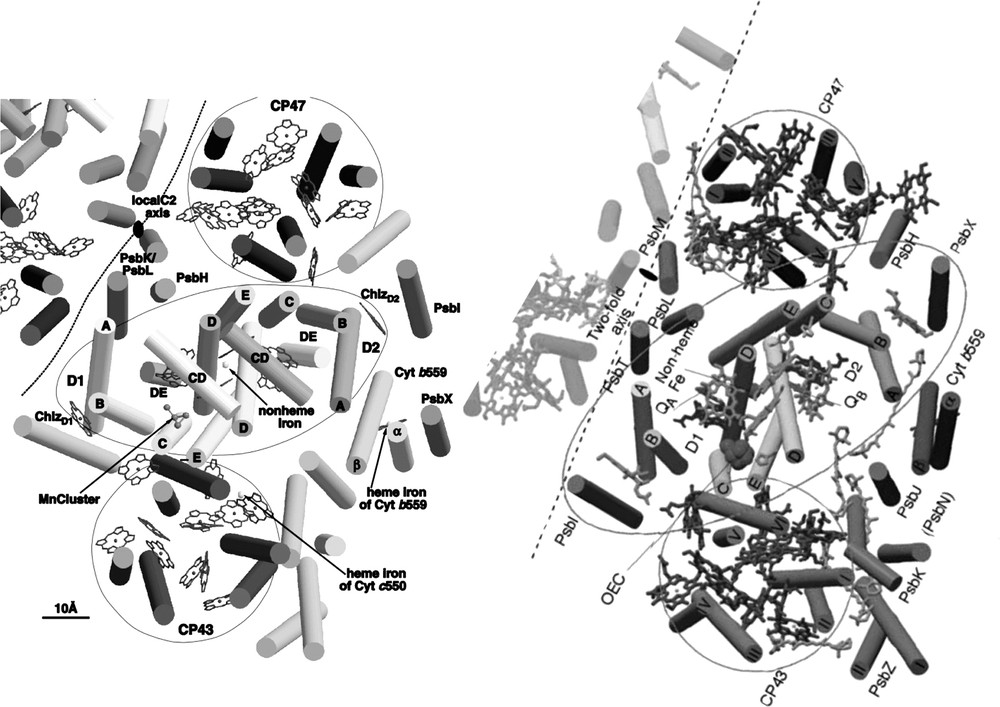
PS II of T. elongatus consists of 17 protein subunits: PsbA to PsbO, PsbU, PsbV and PsbX. Fourteen of these subunits are membrane intrinsic, whereas PsbO, PsbU and PsbV do not contain transmembrane α-helices and are located at the lumenal side of the complex. In the structural model at 3.8-Å resolution [22], the origin of 36 transmembrane helices was analyzed with respect to the localization of the individual subunits, taking into account the available data from biochemical investigations. In the meantime, several more structures from PS II have been published at 3.7–3.2-Å resolution, revealing more details of the structures, including assignments of most of the amino acid side chains and identification of the small membrane intrinsic subunits [24–27]. Fig. 5 shows the comparison of the structural models at 3.8- and 3.5-Å resolution [22,26]. The central core of PS II, which harbors the electron transfer chain, is formed by a cluster of 2 × 5 transmembrane helices, assigned to the protein subunits D1 (PsbA) and D2 (PsbD). The arrangement of these subunits resembles the structure of the L and M subunits of the reaction center of purple bacteria [28,29] and, to a lesser extent, the structure of the C-terminal domain of the large subunits PsaA and PsaB of photosystem I [1], revealing that all photo-reaction centers might have evolved from a common ancestor as previously proposed [30,31].
The central core of D1and D2 is flanked by the antenna proteins CP47 (PsbB) and CP43 (PsbC). Each of these subunits consists of six transmembrane helices. Comparison with the structure of photosystem I reveals that the arrangement of the transmembrane helices of these proteins is very similar to the arrangement of the transmembrane helices in the N-terminal part of the PsaA/PsaB in photosystem I.
Two helices, which are located in close vicinity to helix A of D2, were assigned to the two proteins PsbE and PsbF, constituting the membrane bound cytochrome b559. All other small membrane intrinsic proteins are located peripheral to the central core. The structural assignment of the small subunits is still under debate. The structural model at 3.5 Å [26] contains assignments of all small subunits, whereas the authors of the publication of the structural model at the nominally higher resolution of 3.2-Å state that an unambiguous assignment is not possible at the given resolution and quality of their electron-density map [27].
One helix, present in the 3.8-Å model of PS II, is missing in the 3.5 Å model. The loss may be caused by the higher detergent concentration used for the isolation of PS II in the work of Ferreira et al. [26].
In addition to the transmembrane subunits, PS II contains three extrinsic subunits, which are located at the lumenal side of the core complex: the 33-kDa protein (PsbO), the 12-kDa protein (PsbU) and the cytochrome c550 (PsbV). The latter two subunits are unique to cyanobacteria, whereas PsbO is also present in the photosystem II complex of higher plants. In the structure at 3.8-Å resolution, the main body of PsbO was identified as a β-barrel structure. PsbV was identified by the heme group, which could unambiguously be identified by the high electron density of the Fe ion. The location of PsbU was not resolved at 3.8-Å resolution, but could be identified in the 3.6-Å structure [32,24] as well as in the structures at 3.7-Å [25], 3.5-Å [26] and 3.2-Å [27] resolution. In the improved models, all three extrinsic subunits could be located, as well as most of the loop structures of D1/D2 and CP43 and CP47. The lumenal loop structure of D1 as revealed from the improved structural model at 3.6-Å resolution is shown in Fig. 5 [33,24].
3.2.2 The electron transport chain of PS II
The electron transport chain in PS II consists of four chlorophylls a, two pheophytins, two plastoquinones, one redox active tyrosine TyrZ and the Mn cluster. The position and orientation of the large organic cofactors of the ET chain (i.e. the chlorophylls and the pheophytins) is essentially similar but not identical in all improved structures of PS II. Fig. 6 shows the ET chain as revealed from the 3.6-Å structure of PS II [22,24]. The acceptor side of PS II with the pheophytin, the quinone and the non-heme iron resembles the structural arrangement of the electron transfer chain in reaction centers of purple bacteria. This result is not astonishing, as the physical chemical properties of these cofactors are quite similar in PS II and PbRC. The donor site of the ET chain that differs between the PbRC and PS II will be now discussed in more details.
3.2.3 The primary electron donor: P680
The important differences between PS II and the PbRC are the redox potential of P680, which is more than 1000 mV, and the unique function of water oxidation. One of the most exciting questions is which of the four chlorophylls represent(s) P680.
Four Chl molecules are located in the center of the D1/D2 core. They are arranged in two symmetrically related pairs, P1/P2 and P3 and P4. The chlorophylls named P1 and P2 are oriented parallel to the membrane. The center-to-center distance varies between the different structures: 10 Å in the 3.8-Å structure, 9.56 Å in the 3.7-Å structure, 8.6 Å in the 3.6-Å structure, 8.2 Å in the 3.5-Å structure and 8.3 Å in the 3.2-Å structure. The decrease in distance with increase in resolution may be caused by clearer assignment of the 5/6 ring of the chlorin system at higher resolution. Even with the shortest distance reported to be 8.2 Å, the chlorophylls are more separated from each other in PS II than the chlorophylls in P700 or in the special pair of the PbRC. The longer distance between the two chlorophylls in PS II indicates that the two chlorophylls are not strongly excitonically coupled and therefore may be regarded as chlorophyll monomers. The second pair of chlorophylls is located at a distance of ~10 Å from the first pair of chlorophylls. The plane of the chlorin head group of these chlorophylls is tilted at an angle of 30° to the membrane plane.
The answer as to which of the four chlorophylls may represent P680 may have to be more clearly specified, with respect to whether we want to refer the P680 as the cationic radical P680+•, the triplet state 3P680 or to the excited state P680*. The cationic radical state may be located on P1, because the distance between P680+• and TyrZ has been determined by EPR investigations to be in the range of 7.9 ± 0.2 Å [10] (see also [34,35]), which fits only for the P1 chlorophyll molecule. However, this leads to the question of why the neighboring chlorophylls P2 and P3 may not be oxidized instead of the TyrZ (redox potential 1.0 V) by P1. Taking into account that Pheo–• has an redox potential of 1.4 V, we can assume that all four chlorophylls must have a high redox potential between 1.0 and 1.3 V and may be able to perform the initial charge separation as discussed in [36] and [37]. Therefore, the excited state P680* may be delocalized – at least at room temperature – among all four chlorophylls. The last question regards on which molecule the triplet state of P680 may be localized, which may be responsible for the photodamage of D1. Optical spectroscopy investigations on partially oriented samples provided evidence that the triplet state 3P680 is located on a chlorophyll molecule with an inclination of 30° to the membrane plane [38]. This requirement is fulfilled by P3 and P4, and the location of the triplet on one of the ‘accessory’ chlorophylls has since then been proven by EPR measurements on single PS II crystals [39]. However, the triplet state must be primarily formed from the state P680+• located on P1 by intersystem crossing, therefore P4 can be excluded as a possible candidate and P3 remains as the most probable molecule for the location of 3P680.
The question remains as to what causes the very positive redox potential of P680+•. The binding pocket for P680, as for P700 in PS I, is very hydrophobic, with a histidine providing the 5th ligand to the central Mg2+ ion. However, PS II contains several potentially positively charged amino acids in a vicinity to P680, which may destabilize the cation and increase the redox potential as discussed in [40]. Furthermore, the close proximity of Trp191, which may be π-stacked with D2-His197, which provides the ligand for P2, was discussed as being important for the redox potential shift in [27,41].
3.2.4 The water-oxidizing complex
The most interesting feature of photosystem II is the ability to oxidize water to O2 and 4 H+. Photosystem II performs this reaction by a cluster of four Mn ions, bound to the protein D1. The first structure of PS II at 3.8 Å resolution showed a papaya-shaped structure of the electron density for the Mn cluster, which gave implications for a 3 + 1 organization of the four Mn atoms of the cluster. The subsequent structural models at improved resolution confirmed this principle structural arrangement. However, the arrangements of the Mn atoms differ significantly in the recent structural models, as shown in Fig. 7. Other cofactors that play an important role in the process of water splitting are Ca2+ and Cl–. The location of the Ca2+ has been identified for the first time in the structure of PS II at 3.5 Å resolution [26]. The Ca2+ is clustered together with three Mn atoms, thereby possibly forming a distorted cubane consisting of three Mn and one Ca ion with the fourth Mn being more distal to the distorted cubane. This model of the 4 Mn–Ca cluster is in agreement with EPR and XAFS data.
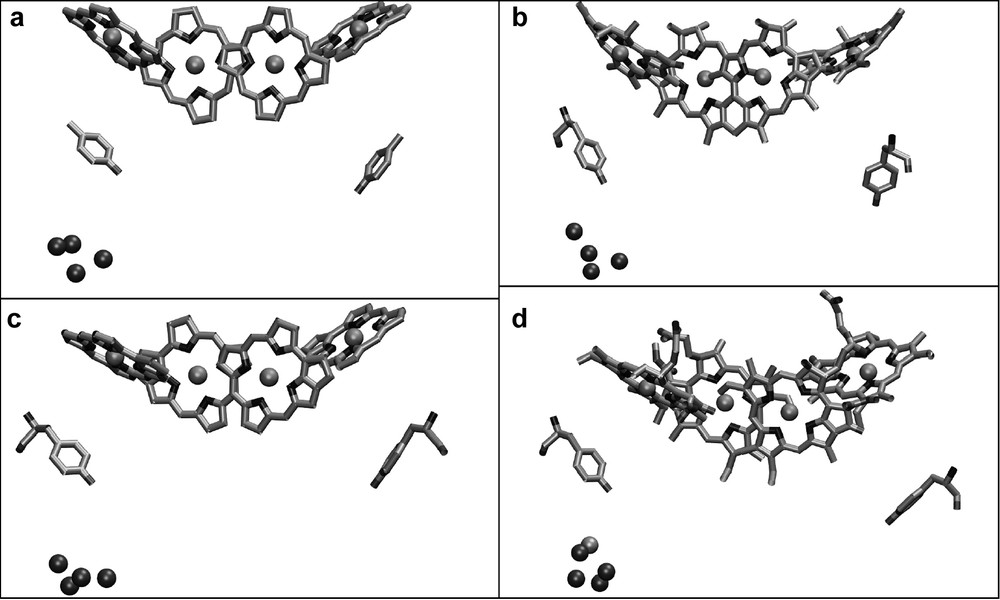
Comparison of the structural models of the Mn cluster in relation to the chlorophylls of the electron transfer chain as revealed from the X-ray structures at (a) 3.8-Å [22], (b) 3.7-Å [25], (c) 3.6-Å [33,24] and (d) 3.5-Å [26] resolutions.
The ligands to the Mn cluster are still under discussion, because they differ in the different X-ray structures and also do not agree in all points with mutagenesis studies. Ferreira et al. have proposed Asp170, Glu189, His332, Glu333, Asp342 of D1 and E354 of CP43 as ligands for the Mn cluster [26]. However, the structure of PS II at 3.2-Å resolution shows a similar but not identical arrangement for the Mn atoms and the ligands. Both structures agree on Asp 170, His 332 and Glu 333 being possible ligands to the Mn cluster. However, Biesiadka et al. see Glu 189 as more distant to the Mn cluster, in a position that may bridge the Mn cluster and TyrZ [27]. In this respect, it is remarkable that Glu189 can be replaced by many other amino acids without affecting oxygen evolution, which leaves the question about the role of Glu 189 completely open. The same is true for CP43-E354, which has been replaced by other amino acids with only a moderate influence on oxygen evolution, whereas the mutation of CP43-R357 (an amino acid which is close, but was not proposed to be a ligand) is lethal. Another controversial question deals with the C-terminus Ala 344. Mutagenesis studies and FTIR experiments propose that it should provide a ligand to the Mn cluster. Indeed, it was supposed to be a ligand in the 3.7-Å structure of PS II [25] but is too distant in both the 3.5- and 3.2-Å structures [26,27].
The final answers to the multitude of questions and uncertainties must be provided by improved X-ray data in combination with spectroscopic and mutagenesis studies. It should also be emphasized that none of the present structural models has a sufficient resolution to identify individual Mn atoms, the bridging oxygens, or the substrate water molecules bound to the cluster, so that the elucidation of the structure of the Mn cluster and the mechanism of water splitting by PS II is still an open field for future discoveries.
3.2.5 The antenna of photosystem II
A total of 42 cofactors have been identified at 3.8-Å resolution: 26 antenna Chl bound to CP47/43, six chlorophylls coordinated by D1/D2 as well as the Mn cluster (consisting of four Mn ions), one non-heme iron, located at the D1/D2 interface on the stromal side of the membrane and the two heme groups of cyt b559 and cytc550, respectively. In the improved structural model at 3.6 Å [32,24], three additional chlorophylls, bound to CP43/CP47 were identified, so that the total amount of chlorophylls was close to the number of 30 chlorophylls assigned at 3.5-Å resolution [26]. All X-ray structural models show very similar locations for the chlorophylls. Fig. 8 compares the chlorophylls assigned in the structural model at 3.6 Å with the chlorophyll arrangement in PS I. The most striking difference between the two is the lack of the central antenna domain in PS II. Whereas more than 50 chlorophylls surround the reaction center domain carrying the electron-transport chain in PS I, the central core in PS II contains only two chlorophylls (ChlZD1 and ChlZD2). This lack of the central antenna domain may be responsible for the lower efficiency of the excitation energy transfer in PS II compared to PS I. The absence of the central antenna domain may be the price PS II has to pay for its ability to use water as an unlimited electron source and being able to undergo the process of photodamage and repair of the D1 protein. If PS II contained a central antenna domain, all central chlorophylls would have also to be replaced with the D1 protein, which would be extremely resource-wasting process.

Comparison of the antenna systems in photosystems I and II.
For clarity, only the chlorophylls are shown. PS II lacks the central domain of chlorophylls that surround the reaction center domain in PS I.
4 Conclusion
The current X-ray structure of PS I and the plethora of experimental data already provide a clear picture of the function of PS I. Nevertheless, there are still a lot of exciting questions open, such as to the role of the lipids, the specific function of the cis-carotenoids, the interaction with peripheral antenna systems and the still open discussion as to which of the electron transfer branches is the active one, the A- or the B-branch. As far as PS II is concerned, the last years brought great improvements towards a better insight into its structure and function. However, many of the central questions still await an answer, such as the mechanism of water splitting, the structure of the oxygen-evolving complex, its ligandation, the role and position of the carotenoids, the function of the small subunits and the repair of PS II. These questions can only be answered by a combination of new discoveries in spectroscopy, molecular biology, model calculations and the determination of a high-resolution structure of PS II in well-defined oxidation states of the Mn cluster.


