1 Introduction
The importance of DNA tests in Forensic Science has been extensively demonstrated. Blood and other fluids are often found at the scene of a crime, and used to obtain information about the possible perpetrators. The fluids can be subjected to DNA typing, and the results are compared to the DNA of suspects and victims. Bloodstains on clothing and surfaces can also be analyzed. DNA sequencing is performed with a variety of chemical tests, and because each person's DNA is different, this can lead to a clear identification of suspects. In many cases, DNA use has led to suspects being found innocent, even though all other evidence pointed to their culpability. Accordingly, innocent persons have been saved from the death penalty or from life imprisonment.
In order to look for new methods of characterizing minute DNA fragments found at the scene of the crime, this work tries to analyze the possibility of alkyllithium reactions with DNA bases and nucleosides. The possibility of DNA reactions with lithium derivatives might contribute to a better understanding of some aspects of DNA identification. Obviously, DNA-containing fluid should be purified and dried beforehand, in order to avoid the simple hydrolysis of the highly sensitive RLi. Beyond this applied interest, the interaction of polar organometallic reagents such as methyllithium with important biomolecules such as nucleotides or DNA fragments has not been studied enough and deserves a preliminary theoretical investigation in order to evaluate new openings in the widely studied chemistry of nucleic acids. The multiplicity of complexation and deprotonation sites in these molecules leads to possible isomers whose relative stability can be predicted using theoretical methods.
The techniques used for the study of these reactions are theoretical, quantum chemical methods, applied to the calculation of the energies of interactions between DNA components (mainly single strand fragments) and methyllithium, taken as a prototype of alkyllithium derivatives.
The interaction of the isolated lithium ion with oxygen or nitrogen atoms of the DNA bases has been the object of a large number of studies in the last years. For instance, the guanine and adenine interaction with monovalent (among which lithium) and bivalent cations was studied by Burda et al. with Hartree–Fock calculations [1a]. Soon after, Lee [1b] performed DFT-B3LYP [2] calculations to the DNA bases' affinity for Li+ and found that the energy of interaction with oxygen is stronger than the one with nitrogen. This method was applied also by Meyer and Suhnel [1c] to the study of the complexes formed by Li, Na or K ions with cyclic base-tetrads. Russo et al. also used the DFT method with the 6-311+G(2df,2p) basis set to evaluate the binding energy and the metalation site of lithium cation for the most stable tautomers of nucleic acid bases [1d]. Zhu and colleagues have probed the interaction of nucleobases with alkaline and alkaline earth metal cations [1e]. Sun and Bu have shown, with the B3LYP method, but with the smaller 6-31+G∗ basis set, that the coupling of Li+ to the guanine–cytosine base pair can strengthen the interaction between guanine and cytosine [1f]. Monajjemi et al. [1g] studied the interactions of Li, Na, Mg and Sr ions with DNA bases, with Hartree–Fock, MP2 and DFT methods. The interaction of a guanine–cytosine pair with a number of metal ions among which Li was studied by Zhao et al. [1h] both at HF and DFT levels and by Reynisson and Steenken [1i] with DFT calculations.
The application of DFT calculations of NMR parameters in order to characterize the monovalent cations' position in guanine quartets was performed by Mourik and Dingley [3a]. Another NMR study of Li+ interaction with nucleosides was reported by Plaush and Sharp [3b].
In addition to the interaction of the DNA bases with the lithium ion, the present study investigates the interaction of some DNA bases, nucleosides and nucleoside pairs with methyllithium. We have retained a set of various DNA fragments that could be representative of diverse experimental situations. For these complexes, the binding can occur in several ways:
- - the binding of the lithium atom in methyllithium with oxygen or nitrogen atoms in the bases, via the formation of a lithium bond;
- - the deprotonation of the bases by methyllithium, which would lead ultimately to the formation of methane and a lithium enolate;
- - the positioning of methyllithium at the phosphate group in two adjacent nucleotides, forming a lithium bond with the oxygens of the phosphate group;
- - the A–T nucleoside pair (double strand) with a methyllithium molecule positioned between the two bases, as an example of double-strand base pair interaction with methyllithium;
All these reactions are likely to be competitive and occur in accord with their own kinetic parameters. This study is not aiming at evaluating their relative speeds; the data we present in the following rather deals with the thermodynamic characteristics of each route.
2 Methods and results
The Spartan [4a] and Titan [4b] programs were used to perform Hartree–Fock and density functional theory (DFT) calculations. The four bases' interaction with Li+ was investigated with the 6-31G∗∗ basis set. Indeed a large number of complexation studies have been published, using the 6-31G, 6-31G∗ and the 6-31G∗∗ basis sets [5]. To investigate the possible role of including diffuse functions, the binding energy of the adenine–Li+ complex was also calculated with the 6-31G+∗ basis set. The result was about 2% smaller than the 6-31G∗∗ result, showing that they are quite similar. The rest of the base–lithium ion complexes were thus studied with the HF/6-31G∗∗ basis set. The initial geometries of the complexes were obtained by positioning the lithium ion systematically in the vicinity of electronegative atoms, such as nitrogen or oxygen. For the sake of brevity, only the most stable isomers are listed in Tables 1 and 2. These structures represent minima since all the eigenvalues are positive and there are no imaginary frequencies. The Zero Point vibrational energy differences for the reaction have been evaluated and found to be in the vicinity of 1 kcal mol−1. Therefore, they are considered negligible and not included in the Tables.
Interaction of the lithium cation with DNA bases at HF/6-31G∗∗ level
| Entry | Complex | Structure | ΔE (kcal mol−1)a |
| 1 | −51.9 | ||
| 2 | Adenine–Li+ | −46.5 | |
| 3 | −54.5 | ||
| 4 | Cytosine–Li+ | −76.0 | |
| 5 | Guanine–Li+ | −78.4 | |
| 6 | Thymine–Li+ | −50.1 | |
| 7 | −51.8 |
a
Binding energies of methyllithium to DNA nucleosides at the 6-31G∗∗ level
| Entry | Complex | Structure | ΔE (kcal.mol−1)a | |
| HF | DFT | |||
| 1 | −25.8 | −29.6 | ||
| 2 | Adenosine–MeLi | −21.6 | −25.5 | |
| 3 | −27.9 | −31.6 | ||
| 4 | Cytidine–MeLi | −32.6 | −32.8 | |
| 5 | Guanosine–MeLi | −29.3 | −30.1 | |
| 6 | Thymidine–MeLi | −27.1 | −26.8 | |
| 7 | −25.8 | −25.3 |
a ΔE = Ecomplex − (Enucleoside + EMeLi).
To calibrate our methodology, we first decided to run calculations on the complexes of DNA bases and an isolated lithium cation. For adenine, three types of complexes have been investigated (Table 1). All the complexes shown and the binding energy values ΔE correspond to geometry optimized at the HF/6-31G∗∗ level. In addition, the complex of cytosine with the lithium ion positioned between the two nitrogens was investigated and found not to represent a minimum in energy. Guanine complexes with Li+ positioned close to two nitrogens were found to be higher in energy by about 40 kcal mol−1.
The 6-31G∗∗ basis set was also used to perform calculations on the complexes obtained by binding methyllithium to each of the four nucleosides. The geometry-optimized complexes are shown in Table 2, which also show the interaction energies, computed either at the HF or DFT (B3LYP) levels using the same 6-31G∗∗ basis set.
Because the above complexes are likely to be transient species en route to a deprotonation, we also investigated this reaction with both the HF/6-31G∗∗ method and the DFT (B3LYP/6-31G∗∗) method. Fig. 1 shows the optimized conformers of the resulting molecules and Table 3 displays the reaction energy of the reaction:
| Base + CH3Li = BaseLi + CH4. |
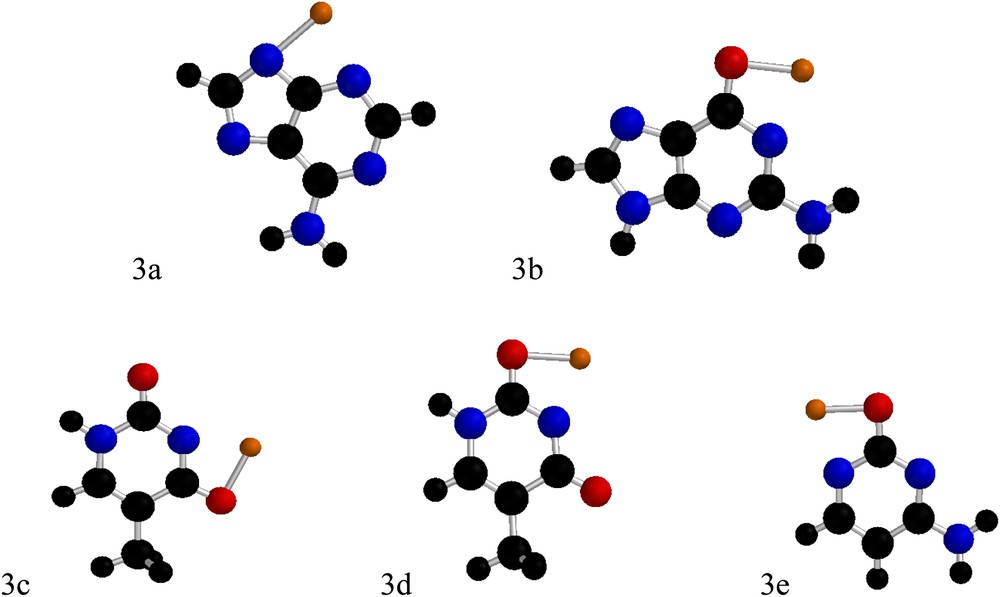
Deprotonated forms of adenine (3a), guanine (3b), thymine (3c and 3d) and cytosine (3e).
Energies of deprotonation of the DNA bases at the 6-31G∗∗ level
| Base | ΔE (kcal mol−1)a | |
| HF | DFT/B3LYP | |
| Adenine 3a | −61.6 | −61.6 |
| Guanine 3b | −72.8 | −70.6 |
| Thymine 3c | −65.3 | −64.3 |
| Thymine 3d | −63.9 | −62.2 |
| Cytosine 3e | −72.0 | −70.3 |
a
The deprotonation of the cyclic NH group in adenine and guanine leads to the corresponding lithium amides (N–Li), while the H abstraction on the NH function of the pyrimidinone in cytosine and thymine affords the lithium 4-aminopyrimidin-2-oxide (O–Li) form. Deprotonated thymine features two regioisomers only (the imide NH being much more acidic than the amide one): the one called 3c which binds lithium to the oxygen near the methyl group and 3d which binds lithium to the other oxygen.
In addition, the deprotonation of the NH2 group in adenine and cytosine by the methyllithium was also considered (leading to the NHLi amide form in 3a1 and 3a2, Fig. 2), as well as the regioisomer of 3e (3e1, Fig. 2). All these molecules proved to be higher in energy than the ones reported above.

Lithium amide forms of adenine (3a1) and cytosine (3e1), and regioisomer of lithiated adenine (3a2) considered in the computations (energy values not reported).
Some single-strand nucleoside pairs–complexes with methyllithium, in which lithium is positioned near the phosphate group, were geometry-optimized with the 6-31G∗∗ basis set. These complexes feature a negative charge on the phosphate group which will, of course, offer an ideal binding site for lithium. The investigated pairs are: A–G, A–T, and T–C and the interaction sites have been restricted to the phosphate appendage. These complexes are shown in Fig. 3 and Table 4 displays their energies and the binding energies of the methyllithium to their phosphate group. In addition, the 6-31G∗∗ basis set was used to calculate the binding energy of methyllithium to an A–T double strand pair, in order to form the complex shown in Fig. 3 (bottom right).

Complex formed between MeLi and the single strand A–G pair (top left), A–T (top right), T–C (bottom left) and double strand A–T pair complex (bottom right).
The binding energies of methyllithium to a single-strand sequence of two nucleotides, at the phosphate group, and to a double-strand A–T pair complex
| Sequence | ΔE (kcal mol−1) HF/6-31G∗∗ a |
| A–G | −41.9 |
| A–T (ss) | −41.5 |
| T–C | −45.7 |
| A–T (ds) | −31.4 |
a E = Ecomplex − (Ebase pair + EMeLi).
3 Discussion
Examining Table 1, one sees that all the DNA bases show a strong affinity for an isolated lithium ion. Guanine and cytosine show the highest binding energy since the ion is positioned in such a way as to bind both to the O atom and to a N atom. For adenine, the structure of entry 3 shows the strongest binding. This is probably due to the chelation of the lithium ion between two nitrogens: the one of the primary amine group (which has rotated by 90 degrees and is not conjugated anymore with the aromatic ring) at a distance of 2.10 Å, and that of the imino-type nitrogen on the five-membered ring (at 1.97 Å). In thymine, the lithium ion can bind to either of the two oxygen atoms and it can be seen that the stronger binding is obtained for the oxygen atom remote from the methyl group (by 1.7 kcal mol−1). The binding energies are in the range previously reported [1].
The first set of molecules investigated for their affinity to methyllithium is the nucleosides (Table 2). Expectedly, the most stable complexes are found when lithium interacts either with the oxygen of a carbonyl or with an aromatic nitrogen. This feature is to be expected from the highly ionic character of the C–Li bond [6,7], a fact which explains that the charged lithium ion (computed between 0.794 and 0.854 e for NBO population in monomeric MeLi at the DFT level) [7] binds via a lithium bond which is mostly electrostatic, especially as shown by NBO calculations [8]. Note that the HF and DFT orders of stability between isomers of a given complex are the same. However, the computed energy of interaction depends on the calculation method: while the formation of guanosine–MeLi complex is more exothermic at the HF level, the adenosine–MeLi one comes first at the DFT level. Overall, cytidine shows the strongest binding followed by adenosine, at least from DFT calculations (entry 3). Among the three different geometries of the adenosine complex, the structure of entries 3 and 1 exhibit the strongest bindings, suggesting a favorable interaction between the NH2 moiety and lithium. Indeed, the Li–N distance (for the N of the amino group) is 2.12 Å in 1, and 2.14 Å in 3.
It can also be seen from Table 2 that the binding energies obtained with the HF/6-31G∗∗ method and with the DFT (B3LYP/6-31G∗∗) method are very close and the trends are the same, as far as the stability of different isomers are concerned.
Another possibility of incorporating lithium in the DNA bases is via their deprotonation with the formation of an NLi bond which can, as expected, rearrange (in the case of guanine, cytosine and thymine) into the isomer featuring an OLi bond. Examining Table 3, which describes the energetics of the deprotonation, one sees that this reaction, which is exothermic, releases the most energy in the case of guanine, followed by cytosine, which affords lithium 4-aminopyrimidin-2-oxy upon NH deprotonation. The two isomers of thymine follow, and adenine exhibits the smallest energy of reaction. However, the difference between thymine and adenine is very small. Thymine exhibits two isomers 3c and 3d. The more stable one is 3c with lithium attached to the oxygen close to the methyl group. With this isomer, the energy of the reaction is higher by about 1.4 kcal mol−1 at the HF/6-31G∗∗calculation level, and by 2.1 kcal mol−1 using the DFT method. For adenine, the deprotonation takes place on the five-membered cyclic NH group. Note that the overall order of exothermicity is conserved.
In all these compounds, the lithium atom is positioned (via geometry optimization) in such a way as to be in bonding distance of both the atoms to which it is attached, and of a near-by nitrogen. The Li–O bonds are 1.80 Å for guanine, 1.81 for cytosine, 1.87 for 3a and 1.89 for 3b. The Li–N distances are from 1.93 to 2.00 Å (at the HF/6-31G∗∗ computational level).
Let us finally discuss the case of the base pairs. These calculations were performed with the HF/6-31G∗∗ method. Methyllithium, as seen in Table 4, binds to the phosphate group. The highest binding energy occurs for the T–C sequence, followed by the A–G and then A–T sequence. When the complex is formed, the Li–C bond in MeLi is elongated by about 0.1 Å. The distance between Li and the oxygens of the phosphate is 1.7 Å in the A–T complex, 1.9 Å in the T–C complex and 2.1 Å in the A–G complex. The distance between Li and the oxygens of the phosphate group is around 1.9 Å.
4 Conclusion
The above results suggest that the reaction of a strongly polar organometallic reagent such as methyllithium with the DNA bases and their derivatives is likely to afford metastable complexes (local minima) probably intermediates in the reaction of deprotonation of one of the acidic NH protons. It is to be believed that the global minima will follow the deprotonation of one of acidic NH protons and formation of a C–O–Li or C–N–Li group. It might be concluded that the methyllithium primary interaction with DNA fragments will proceed via the incoporation of the lithium atom in the bases, following deprotonation. All the complexes investigated are minima (local or global) since there are no negative eigenvalues or imaginary frequencies. In the case of the isolated Li+ cation, the interaction occurs with two of the polar groups in the molecule. Note that the base–Li+ binding energies obtained (Table 1) are approximately as high as the deprotonation energies shown in Table 3. It is probable that the gas-phase binding energies are larger than the binding energies occurring in solution. Since water cannot be used as a solvent, other solvents will be investigated in future studies.
These biomolecules' involvement in crime scene investigations is taking increasing importance. Our results might help elucidate the DNA fragments' characterization through for instance 6Li [9] or 7Li [10] NMR analysis on the resulting products. In addition to experimental data concerning these entities, our calculations can help to characterize complexes of DNA fragments with methyllithium, leading to possible applications of small DNA fragments' identification, as relevant to crime elucidation. Indeed, one of the complex problems related to the study of DNA fragments found at a crime scene is the dating of the DNA from different tissues or dried fluids. Thus, the degradation of the DNA molecules with time could provide information about the time elapsed between the commission of the crime and the finding of the DNA fragments. Experiments (NMR, IR) that would characterize the change of the nature of the DNA base complexes with methyllithium as dependent on the time elapsed, might contribute to clarify this issue. Let us finally underline once again that all these calculations presume a non-aqueous medium. In the presence of water, methyllithium would, of course, decompose before any other reaction.
Acknowledgements
We want to thank Dr. L. Kobilinsky for useful discussions.



Vous devez vous connecter pour continuer.
S'authentifier