1 Introduction
1.1 Multiple meanings for “synthetic biology”
Many languages have words and phrases, called contranyms, that have two nearly opposite meanings. For example, a “citation” from Harvard University is good, but a “citation” from the Harvard University police is bad. If you run “fast”, you are moving at great speed; if you hold “fast”, you are not moving at all.
“Synthetic biology” is a contranym. In a version popular today in some engineering communities, it seeks to use natural parts of biological systems (such as DNA fragments and protein “biobricks”) to create assemblies that do things that are not done by natural biology (such as digital computation or manufacture of a speciality chemical). Here, engineers hope that the performance of molecular parts drawn from living systems can be standardized, allowing them to be mixed and matched to give predictable outcomes, just as transistors are standardized allowing engineers to mix and match them to give integrated circuits with predictable performance. For this, the whole must be the sum of its parts.
Among chemists, “synthetic biology” means the opposite. Chemist's “synthetic biology” seeks to use unnatural molecular parts to do things that are done by natural biology. Chemists believe that if they can reproduce biological behavior without making an exact molecular replica of a natural living system, then they have demonstrated an understanding of the intimate connection between molecular structure and biological behavior. If taken to its limit, this synthesis would provide a chemical understanding of life. Although central to chemistry, this research paradigm was perhaps best expressed by a physicist, Richard Feynman, in the phrase: “What I cannot create, I do not understand.” [1].
Waclaw Szybalski had yet a different meaning in mind when he coined the term “synthetic biology” in 1974 [2]. Szybalski noted that recombinant DNA technology would soon allow the construction of new cells with rearranged genetic material. He realized that this deliberate synthesis of new forms of life provided a way to test hypotheses about how the rearranged material contributed to the function of natural cells.
Szybalski had the experience of chemistry in mind when he coined the term. In 1974, Structure Theory in chemistry was the most powerful theory in science. It became so largely because chemistry possessed technology that allowed chemists to synthesize new chemical matter to study. This supports powerful processes for testing hypotheses and models, power that Szybalski saw that biotechnology was delivering to biology. Such a power was (and remains) unavailable to (for example) astronomy, planetary science, and social science.
In 1974, “synthetic organic chemistry” had already penetrated into biology. For example, in the previous decade, “biomimetic chemists” had created small designed molecules that reproduced the elementary behavior of biomolecules, such as their ability to bind to ligands or to catalyze reactions. Jean-Marie Lehn in Strasbourg shared a Nobel Prize for his work developing molecules able to do exactly this. One of his signature structures from the 1960s is shown in Fig. 1.

Synthesis in biology was first used to help understand the connection between biomolecular structure and behavior by making unnatural molecules that bind to small molecules. This synthetic receptor was created by Jean-Marie Lehn and his colleagues to mimic the ability of natural receptors to bind to small cationic ligands, and was cited in his Nobel Prize lecture.
1.2 Challenges in contemporary synthetic biology
Today, the chemist's vision for synthetic biology goes further. The hope is that molecular design supported by Structure Theory will yield unnatural molecular species able to mimic not just binding and catalysis of specific biomolecules, but also the highest kinds of biological behavior, including macroscopic self-assembly, replication, adaptation, and evolution. Any theory that enables such design will have demonstrated an ability to account for these features of “life”, especially if chemists can make a totally synthetic version of life without exactly reproducing the chemistry of a natural terran organism. With the term “synthetic biology” now in jeopardy by “trademark creep”, it might be appropriate to coin a new term to describe this process, perhaps “Artificial Biology”, although computer engineers have already used this term to mean something different.
Given these nearly opposite uses of the same term, spectators are naturally puzzled. I am often asked by reporters about the emerging use of “synthetic biology” in the engineering sense: “What's the fuss? Isn’t synthetic biology just more ‘Flavr Savr’® tomatoes?” The question is raised in analogous form by molecular biologists who see in synthetic biology “contests”, which attract student participation worldwide, nothing more (and nothing less) than the cloning that has been done since the 1970s. At worst, it illustrates the aphorism that “the difference between men and boys is the price of their toys”.
Nor do molecular biologists attempting to understand life entirely understand the hullabaloo over the (difficult to repeat) use of DNA hybridization and ligation to compute a solution to the “traveling salesman problem” [3]. There is no obvious reason to do this kind of digital computation with DNA. After all, the rate at which DNA molecules hybridize in solution is limited by the rate constant for molecular diffusion, about 108 M−1sec−1. In layman's language, this means that the half-life with which a DNA molecule finds its complement cannot be faster than ca. 0.01 sec when the complement is at a high concentration of one micromolar. In practice, the rate is much slower. Contrast this with the limit on the rate at which semiconductors compute: the speed of light in the conducting material. At 3 × 1010 cm/sec, communication across a meter of space is ∼0.000000003 sec. Even with the possibility of improved parallelism with DNA computation, there is no contest.
Francis Collins, the newly appointed director of the National Institutes of Health in the United States, captured a similar sentiment. Collins is reported to have mused about the “new” field of synthetic biology as applied to virus synthesis: “This was completely a no-brainer. I think a lot of people thought, ‘Well, what's the big deal? Why is that so exciting?”’ [4].
1.3 This is not the first time that biology has been declared to be engineerable
Salesmanship accompanying some discussions today of “synthetic biology” has also engendered a degree of cynicism [5]. Those whose professional lives started before the age of the Internet remember more than one time where biology, it was claimed, had at last entered the realm of engineering. It was not so then and, in the broadest vision put forward by the engineering community, it is not so now.
For example, a quarter century ago, Science published an article entitled “Protein Engineering” by Kevin Ulmer, Director of Exploratory Research at GeneX, a biotechnology company [6]. Science has not placed papers published before 1997 on the Internet; one goes to a library to get this paper. Other than that, the 1983 paper is both in substance and form like the breathless reporting in today's popular science magazines covering efforts to make and use biobricks.
In 1983, Ulmer said that the goal of this new engineering biology was to “control in a predictable fashion” the properties of proteins to be building blocks in industrial processes. This new era of engineering would set aside “random mutagenesis techniques” in favor of a “direct approach to protein modification”. Ulmer referred to protein domains encoded by exons and the use of repressors with altered enzymes to assemble new regulatory pathways.
Ulmer's 1983 vision failed. GeneX is no longer in business. Twenty-five years later, we are still struggling to engineer the behavior of individual proteins.
The 1980s engineering vision failed for reasons discussed in a 1987 review by Jeremy Knowles, then Dean of Harvard College, entitled Tinkering with Enzymes [7]. Knowles, a chemist, understood that “scale” matters. Molecules, one to one-tenth nanometer in size, behave differently from transistors, even transistors existing at the one to one-tenth micrometer scale. This creates difficulties in transferring microengineering concepts to molecular engineering. The same is true in the next jump downwards in scale; molecules at one to one-tenth nanometer scale behave differently from quantum species operating on the pico- or femtometer distance scales.
Knowles's “Tinkering” comments are apt even today. Referencing Ulmer's paper, Knowles dryly wondered whether the engineering vision was not, perhaps, a bit “starry-eyed”. He acknowledged that “gee whiz” experiments that put things together to “see-what-happens” could aid in understanding. But he made the point that is still true, and which is a theme of this lecture: Nothing of value comes unless the tinkering is followed by studies of what happened. Especially if the synthetic effort fails. Absent that, modern synthetic biology, at the molecular, DNA, protein, or cell level, will be “tinkering” without consequence.
Analysis of failure is generally less enthusing (and more laborious) than the initial design, as evident to any observer of synthetic biology “contests”. Indeed, the analysis of failure requires discipline, a discipline that is difficult to teach. Thus, it helps to remember the dictum: It is just as hard to solve an unimportant problem as an important one. One is more likely to analyze a failure to the depth needed to learn from that failure if the goal is felt to be very important.
We understand much more now about the behavior of molecules, biological and otherwise, than we understood a quarter century ago because tinkers studied their failures. Accordingly, the ball has been moved, from Lehn-like small molecules to proteins to synthetic genes, protein assemblies, cells and assemblies of cells. We are still doing what might be called “Tinkering Biology”, but we are doing it farther down the field. This term will be appropriate until failures in synthetic biology are commonly examined to understand what went wrong.
1.4 What do opposite meanings of “synthetic biology” have in common?
One lesson in particular might be learned from these “pre-Internet” failures in synthetic biology. Synthesis is a research strategy, not a field [8,9]. Synthesis sets forth a grand challenge: “Create an artificial chemical system capable of Darwinian evolution.” Or: “Create a set of DNA bricks that can be assembled to form an adding machine.” Or: “Rearrange a set of regulatory elements to make a cell that detects nerve gas.” Or: “Assemble enzyme catalysts taken from a variety of organisms to generate a pathway to make an unnatural chemical that is part of an antimalarial drug.” Attempting to meet this challenge, scientists and engineers must cross uncharted territory where they must encounter and solve unscripted problems guided by theory. If their guiding theory is adequate, the synthesis works. If it is not, the synthesis fails.
This exercise is different from observation, analysis, and probing, other strategies used in science. Here, as often as not, observations are often either discarded or rationalized away when they contradict a (treasured) theory. We see this also in computational modeling using numerical simulations. “Modeling”, it is said “is doomed to succeed.” If a model does not give a desired answer, it is tweaked until it does.
Selection of data to get the “right” answer has a long tradition in biology. A well-known example is Gregor Mendel, who evidently stopped counting round and wrinkled peas when the “correct” ratio (which is 3:1) was reached. Objective observations have an uncanny ability to confirm a desired theory, even if the theory is wrong.
Self-deception is far more difficult when doing synthesis. If, as happened with the Mars Climate Orbiter, the guidance software is metric and the guidance hardware is English, one can ignore the incongruent observations arising from a false theory (as was done) as the craft was in transit to Mars. But when the rocket gets to Mars, if the theory is wrong, the rocket crashes (and it did).
For this reason, synthesis as a research strategy can drive discovery and paradigm shift in ways that observation and analysis cannot. Indeed, I have gotten in the habit of saying that this is one way to distinguish science from non-science: Science is a human intellectual activity that incorporates a mechanism to avoid self-deception [9]. Synthesis provides such a mechanism.
1.5 The value of failure, and the analysis of failure
The failures encountered as modern synthetic biologists attempt to rearrange atoms to create artificial genetic species [10], regulatory elements to give synthetic circuits [11], or enzymes to give synthetic pathways [12] carry a clear message: we need to learn more about the behavior of physical matter on the “one to one-tenth” nanometer scale. This is not a declaration of defeat. Rather, it is a challenge, one that begins by recognizing that our guiding theory is still inadequate to hand biology over to engineers.
Nowhere is the value of failure set within a synthetic challenge more evident than within recent efforts of Craig Venter, Hamilton Smith, and others to construct a cell where all of its constituent genes come from elsewhere [13,14]. In a real sense, this grand challenge is the apotheosis of the 1970s version of bioengineering. More Flavr Savr® tomatoes, but now thousands of times repeated.
On paper, the challenge appeared to be simple enough. Since the time of Szybalski, scientists had been able to move a single natural gene from one organism to another. Scientists had long been able to move two natural genes into, from two other organisms, a different organism. By a kind of argument that corrupts the idea behind mathematical induction (if one can do n genes, and if one can do n + 1 genes, one should be able to do any number of genes), it seemed that simple iteration would allow scientists to get all genes in an organism from somewhere else.
When these scientists set out to do this, they had high hopes. I had dinner with Hamilton Smith, in October 2006. They were just “6 months away” from getting a synthetic cell constructed in this way. We met again 18 months later at Janelia Farms; they were still 6 months away. Three years later, the final announcement of success [14] was made just as this article was going to press. The estimated price tag of $40 million (US) shows just how great this challenge was?
The difficulties arose because the operating theory used to guide this synthesis was missing something. In the case of the synthetic cell, Venter noted that a single nucleotide missing in a critical gene prevented success for many months. Again, this is not a reason for despair. The purpose of synthesis is to bring those missing “somethings” to light. As they pursued their grand challenge, Smith, Venter, and their colleagues had to solve unscripted problems, driving knowledge in ways impossible through analysis and observation alone.
Here as always, problem selection is important. Selecting problems at the limits of the possible is a poorly understood art. Further, those selections change over time. For example, in the 1960s, it was a sufficient as a challenge to try to develop organic molecules that would bind to small molecules, such as the synthetic receptors that earned Jean-Marie Lehn his Nobel Prize (Fig. 1).
Many challenges undertaken today under the synthetic biology paradigm seek to create “gee whiz” toys. Venter's challenge was certainly not this. The most useful challenges are those that are most likely to generate the most consequential pursuits. These are the challenges just at the limits of the do-able, and perhaps just a bit farther. As Medewar said, science is the “art of the soluble” [15]. However, the selection of a synthetic challenge also reflects choices personal to scientists in a laboratory. This lecture is about the choices made in my laboratory by my co-workers and me in a kind of synthetic biology that is more oriented towards chemistry.
2 Attempting to synthesize an artificial genetic system
Our efforts in synthetic biology began in the mid-1980s, immediately after my group synthesized the first gene encoding an enzyme, and the first synthetic gene of any kind to be designed with computer assistance [16], following a similar gene encoding interferon [17]. Our next “grand challenge” was to synthesize unnatural chemical systems that do genetics, catalysis, and the higher order functions in life, including reproduction, adaptation, and evolution. The first book in synthetic biology, originally entitled Redesigning Life, appeared in 1987 [18].
It took 20 years to create an artificial chemical system capable of Darwinian evolution, two decades of more failures than successes. Again, no problem. This is what synthesis is for. Let me highlight some of the parts of our work to illustrate what synthetic efforts directed towards this challenge taught us. More details are available in a new book Life, the Universe, and the Scientific Method (http://www.ffame.org) [19].
Our selection of this particular challenge focused on a broad question: how might we use synthesis to develop a better understanding of the concept of “life”? This question is, of course, at the abstract core of biology. But it also has practical implications. For example, NASA and ESA send missions to Mars, Titan and elsewhere, looking for life in environments that are more or less like Earth. On Mars, environments are more like Earth, as liquid water most likely lies beneath the Martian surface. On Titan, conditions are less like Earth. The most abundant matter on Titan's surface that might serve as a biosolvent is liquid methane at 94 K (−179 °C); the liquid water beneath the surface may exist as water-ammonia eutectics. Nevertheless, organic species are abundant on Titan. Thus, as many have argued, if life is an intrinsic property of organic species in complex mixtures [20], then Titan should hold life.
Accordingly, my laboratory set up a set of programs in the 1980s to pursue four approaches towards understanding the concept of “life”. These are illustrated in Fig. 2.
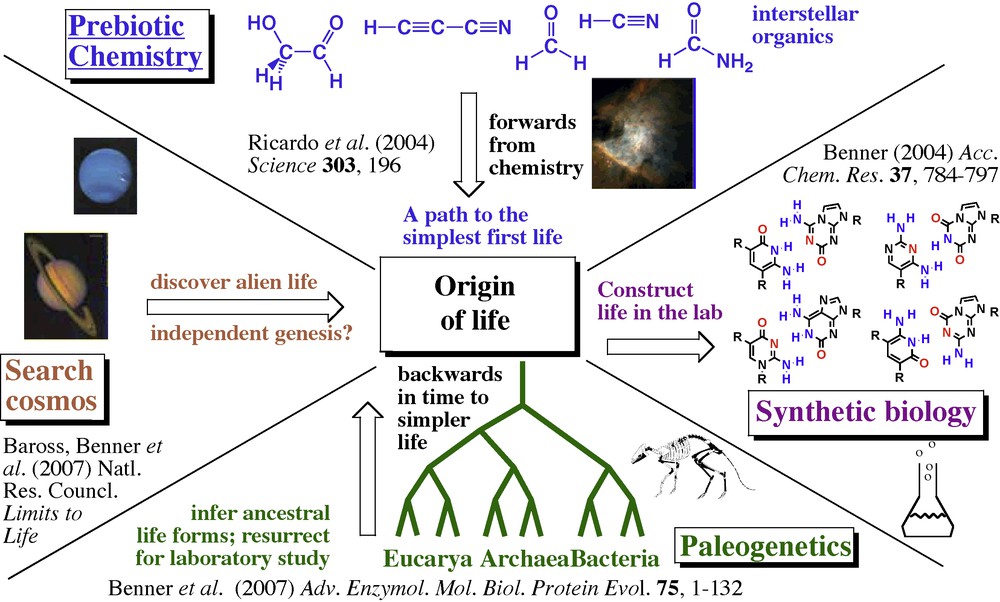
Four approaches to understand life as a universal.
Any definition of “life” must be embedded within a “theory of life” [9,21]. One such theory is captured by a “NASA definition” that “life is a self-sustaining chemical system capable of Darwinian evolution” [22]. This definition-theory captures the opinion of its creators about what is possible within biomolecular reality. It excludes, for example, non-chemical and Lamarckian systems from our concept of “life” [23]. Should we encounter them (and many science fiction stories describe them in various forms), we would be forced to concede that our definition-theory of life is wrong.
The NASA definition-theory of life offers a clear direction for exploratory synthesis. If this theory describing life were so simple, then a target for “synthetic biology” would be an artificial chemical system capable of Darwinian evolution. If the NASA definition-theory of life is on point, this artificial system should be able to recreate all of the properties that we value in life. To manage issues of ethics and hazards, we might not seek to make our system self-sustaining, unlike (for example) the synthetic constructions sought by Venter, Smith, and their colleagues.
Like the Venter-Smith team two decades later, we also had high hopes when we set out. After all, the existing theory at that time, constructed at the molecular level, seemed to associate Darwinian evolution with some quite simple molecular structures, even simpler than those required to assemble an artificial cell from natural genes taken from elsewhere.
2.1 Synthesizing artificial genetic systems
Consider, for example, the double helix structure of DNA, the molecule at the center of natural Darwinian evolution, modeled by James Watson and Francis Crick in their epic 1953 paper. In the double helix, two DNA strands are aligned in an antiparallel fashion. The strands are held together by nucleobase pairing that follows simple rules: A pairs with T and G pairs with C. Behind the double helical structure lay two simple rules for molecular complementarity, based in molecules described at atomic resolution. The first rule, size complementarity, pairs large purines with small pyrimidines. The second, hydrogen bonding complementarity, pairs hydrogen bond donors from one nucleobase with hydrogen bond acceptors from the other (Fig. 3).
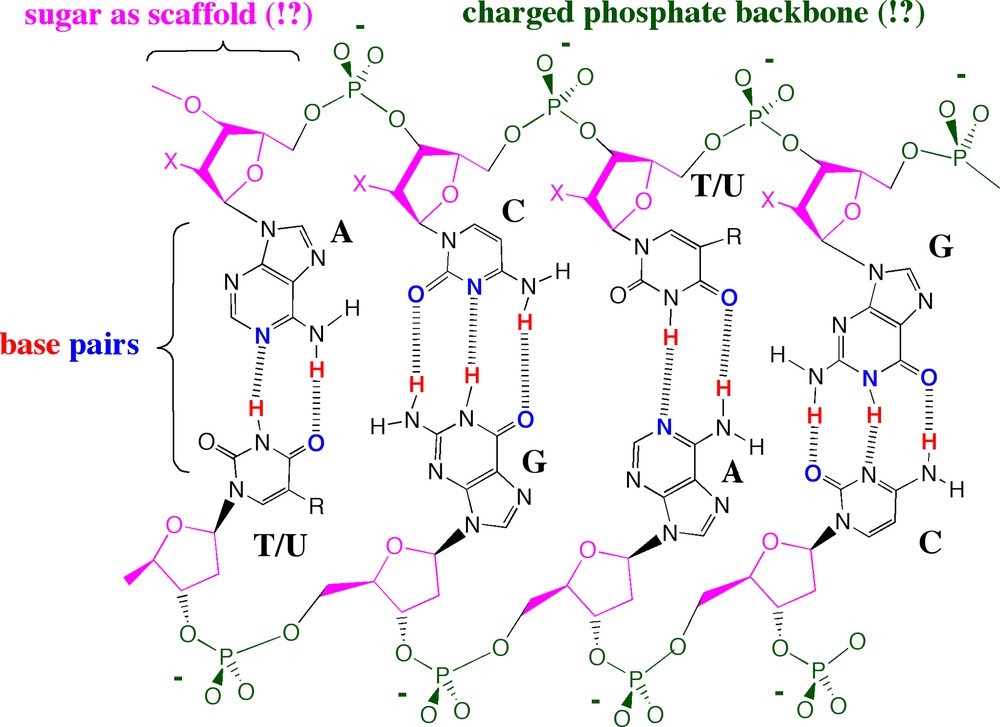
The rules governing the molecular recognition and self-assembly of DNA duplexes are so simple that many are tempted to believe that molecular recognition in chemistry is, in general, similarly simple. Hence, various individuals seek “codes” for protein folding or drug binding.
In the first generation model for the double helix, the nucleobase pairs were central. In contrast, the backbone “bricks”, made of alternating sugar and negatively charged phosphate groups, were viewed as being largely incidental to the molecular recognition event at the center of natural genetics and Darwinian evolution.
2.2 Failure changing the sugar
If this simple “first generation” model for the double helix were correct and complete, then we should be able to synthesize a different molecular system with different sugars and/or phosphates (but the same nucleobases) to get an unnatural synthetic system that could mimic the molecular recognition displayed by natural DNA and RNA. We might even be able to get this artificial genetic system to have children and, possibly, evolve.
In our efforts, the only ones about which I can speak authoritatively, much failure ensued. For example, we decided to replace the ribose sugars by flexible glycerol units to give a “flexible” kind of synthetic DNA (Fig. 4). This followed a suggestion of Joyce, Schwartz, Miller and Orgel [24], who had noted some of the difficulties in identifying processes that, on Earth before biology, might have generated ribose and 2’-deoxyribose, the “R” and the “D” in RNA and DNA respectively. Glycerol, from a prebiotic perspective, was certainly more accessible to a pre-life Earth; it is a major component of organic material delivered to Earth today by meteorites [25].
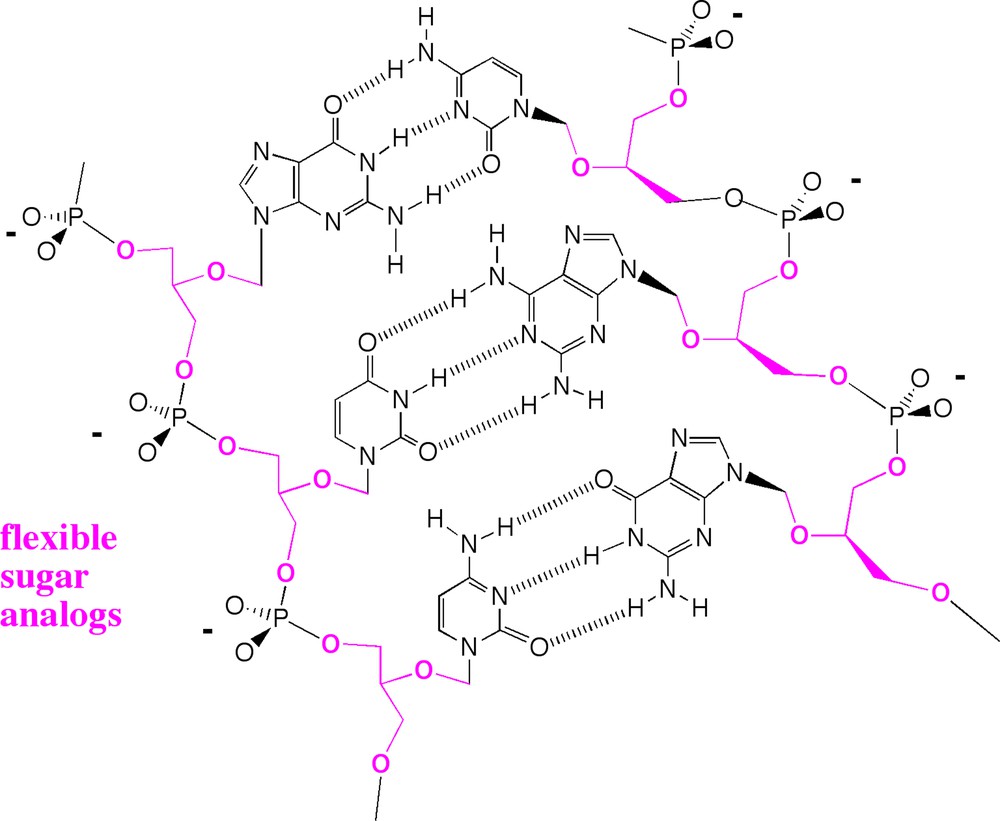
The failure of these flexible glycerol DNA molecules led us to re-evaluate our view of the role of sugars in double helix formation.
Unfortunately, the system failed to deliver quality rule-based recognition. As summarized in Table 1, synthetic molecules that replaced one ribose by a glycerol all bound less tightly to their complement. Putting in two flexible glycerols lowered the melting temperatures of the duplexes even more. Faced with this failure, we went further, synthesizing DNA analogs where all of the 2’-deoxyribose units were replaced by glycerol units. Making the molecule entirely from glycerols destroyed all of the molecular recognition needed for DNA-like genetics.
Melting temperatures for flexible glycerol synthetic DNA.
| CTTTTTTTG | 40° | CAAATAAAG | 37° |
| GAAAAAAAC | GTTTATTTC | ||
| CTTTtTTTG | 25° | CAAAtAAAG | 25° |
| GAAAAAAAC | GTTTATTTC | ||
| CTTtTtTTG | 13° | CAAtAtAAG | 12° |
| GAAAAAAAC | GTTATATTC | ||
| CTTttTTTG | 11° | CAAttAAAG | 11° |
| GAAAAAAAC | GTTAATTTC | ||
| CTTTTTTTG | 21° | CTTTtTTTG | 12° |
| GAAAGAAAC | GTTTGTTTC | ||
| CTTTTTTTTTTTG | 55° | CtttttttttttG | < 0° |
| GAAAAAAAAAAAC | GAAAAAAAAAAAC |
This failure taught us the inadequacy of our then-existing theory to account for genetics. Two hydrogen bonds joining the nucleobase pairs were simply not enough to hold together two strands where the ribose was replaced by synthetic glycerol. Conversely, these experiments showed that the sugars were not entirely incidental to the molecular recognition event.
When three hydrogen bonds held the nucleobase pairs together, things work better. Further, with a new-found appreciation of the contribution of the sugar to the ability of DNA to support Darwinian evolution, synthesis went further. Smaller carbohydrates and carbohydrates whose conformation was locked were synthesized and found to work better than standard DNA, at least by some metrics [26,27]. These themes were further developed by the synthesis of more backbone-modified DNA species by many luminaries in modern synthetic chemistry, including Albert Eschenmoser [28], Piet Herdewijn [29], and Christian Leumann [30].
Thus, a theory that taught that the backbone sugars of DNA were incidental to Darwinian evolution failed to support a synthetic endeavor. This failure advanced the theory. The synthesis of unnatural genetic systems taught us something about natural genetic systems. This drove the synthesis of more unnatural systems that replaced failure by success.
Without synthesis, this part of the first generation theory for the double helix had remained largely unchallenged in the three decades since it was first adumbrated in 1953. It had appeared as “dogma” (Francis Crick's word) in textbooks and television series. Some of these said that “RNA is the same as DNA, except that each sugar has an additional –OH group” (a statement that, from any chemical perspective, is ignorant on its face). These facts all support the notion that without the efforts of synthetic biology, this fascinating feature of the molecule behind Darwinian evolution would never have been recognized. Synthesis drove discovery and paradigm change in ways that analysis cannot. And this came about only because failure was analyzed and pursued.
2.3 Failure changing the phosphates
Failure was also encountered when we attempted to replace the charged phosphates in the backbone of DNA by a linker that had about the same size as phosphate, but that lacked charges. The phosphate linkers were also viewed in the first generation Watson-Crick theory as being largely incidental to the molecular recognition that is central to genetics and Darwinian evolution.
In fact, the repeating negative charges carried by the phosphate groups in the DNA backbone appeared to be downright undesirable. The repeating charges on the phosphate linkages prevented DNA from getting into cells. The charged phosphate linkers were sites of nuclease attack. The repulsion between two negatively charged backbones of two DNA strands seemed to weaken undesirably their association to form a double helix. DNA molecules without the negative charges in their backbone were expected to form better duplexes, an example of the “God made a mistake” theme.
If, it was thought, we could get rid of the charges without disrupting the rules for Watson-Crick pairing (A pairs with T, G pairs with C), we might be able to create a new class of therapeutic molecules with an entirely new mechanism for biological activity. These were called “antisense drugs” [31]. The idea was simple. If we could synthesize an uncharged analog of DNA that could enter a cell by passive diffusion, it would survive degradation by nuclease attack. If the charges were indeed incidental to genetics, this neutral synthetic DNA analog would still bind to complementary DNA molecules inside a cell following Watson-Crick rules. The antisense DNA analog would therefore target, with sequence specificity, only the unwanted DNA, perhaps from a virus or a mutated cancer gene. Antisense DNA might be a magic bullet for diseases associated with undesired DNA or RNA.
Following this theory, Zhen Huang, Christian Schneider, Clemens Richert, and others in my group synthesized an uncharged unnatural DNA-like molecule that replaced the anionic phosphate diester linker in natural DNA and RNA with uncharged dimethylenesulfone linkers (Fig. 5). This gave DNA and RNA analogs that have roughly the same geometry as the natural molecules [32,33]. Indeed, Martin Egli solved a crystal structure of a short GSO2C dinucleotide duplex. He found that the uncharged duplex was held together by G:C and C:G pairs in a minihelix just like its RNA analog, whose crystal structure had been solved by Alex Rich two decades earlier [34].
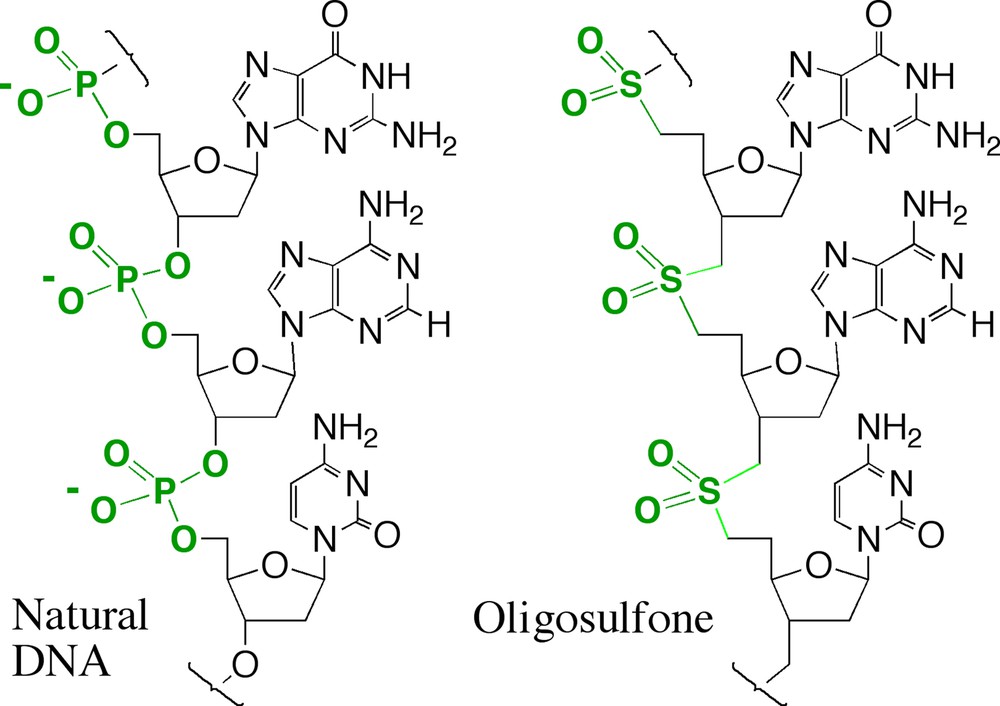
Replacing phosphates (–PO2- units, each having a negative charge, left) in DNA by dimethylenesulfone linkers (the –SO2- units, right, each lacking a negative charge) gave an uncharged analog of DNA. The uncharged analog of RNA was also synthesized.
This appeared to validate the first generation Watson-Crick theory for the double helix. It appeared that one could replace the charged phosphate linkers with uncharged linkers of approximately the same shape, and still form G:C and C:G pairs.
One theme already mentioned is that serious synthetic biologists do not neglect detailed analysis when a synthesis fails. The contranym theme is that one should extend the challenge when the synthesis appears to succeed. Success means that one has erred a bit on the safe side in selecting a challenge. To be consequential in driving discovery and paradigm change, if a theory seems to work, the challenge should be deepened until the theory fails.
Accordingly, we synthesized longer DNA and RNA analogs having more sulfone linkers. Instead of molecules with just one uncharged linker, we made molecules with two uncharged sulfone linkers to see how they worked. We then made molecules with three, five, and then seven uncharged sulfone linking bricks.
It was not long before the theory that we were using to guide the synthesis broke down. Longer oligosulfones folded on themselves [35]. Folding prevented them from pairing with any second strand, even one that was perfectly complementary in the Watson-Crick sense of the term. This failure led to a thought that should have been obvious, but was not in our culture (we too had been trained to view the DNA double helix as an unchallengeably elegant structure): pairing between two strands requires that neither strand fold on itself.
Another failure was then encountered. Different oligosulfones differing by only one nucleobase in their structure were found to display different levels of solubility, aggregation, folding, and chemical reactivity [36]. This prompted another thought that, in retrospect, should have been obvious. To support Darwinian evolution, a genetic molecule must have features that allow it to change its detailed structure, the details that encode genetic information. However, the changes must be possible without changing the overall properties of the system. In particular, the changes in structure that correspond to changes in genetic information cannot change the rules by which the genetic molecules template the formation of their descendents. Changes do not do this in DNA and (in general) RNA. As we learned by synthesis, they do so in oligosulfones.
These results further drove the development of a second-generation model for the DNA double helix and the relation between its structure and Darwinian evolution. In this model, the phosphate linkers and the repeating backbone charge become quite important for four reasons.
First and trivially, a polyanion is likely to be soluble in water. This was appreciated by Watson and Crick already in 1953, certainly more than Linus Pauling. Pauling had proposed an incorrect model for DNA where the phosphates did not point out into solvent, but rather (and paradoxically given their negative charges) interacted with each other [37]. When Watson and Crick first learned about the structure for DNA assemblies that Pauling was proposing, this feature immediately let them conclude that Pauling's model must be wrong.
Less trivially, the repeating charges in the backbone of natural polyanionic DNA repel each other. Within a strand, this repulsion helps keep DNA strands from folding on themselves. A polyanion is more likely to adopt an extended conformation suitable for templating than a neutral polymer, which is more likely to fold. As “not folding” is a property needed for a strand to bind to its complement, the repeating charges were proposed in the second-generation model to be important for the ability of DNA to support Darwinian evolution for this reason, as well as for solubility reasons.
The anion-anion repulsion between phosphates on two different strands is also important. When two strands approach each other, the repulsion forces inter-strand interactions away from the backbone. This drives the contact between two strands to occur at the Watson-Crick edge of the nucleobases (Fig. 6). Without the polyanionic backbone, inter-strand contacts can be anywhere [38]. Thus, the second-generation model views as naive the assumption that this repulsion is bad. In fact, the repulsion moderates and controls the natural propensity of biomolecules to associate with other biomolecules, and directs in DNA that association to the part of the molecule where information is contained, the Watson-Crick edges of the nucleobases.

The repeating backbone anion drives the interaction between two strands as far from the backbone as possible. This guides strand-strand interactions, and forms the basis for Watson-Crick pairing rules.
In the light cast by failure in a synthetic effort, the inter-strand repulsion between two strands that both have repeating charges on their backbones is also seen to be important for pairing rules essential for Darwinian evolution. Without the repulsion from two backbones, both negatively charged, base pairing would not occur at the site where hydrogen bonding was needed. It would occur at other sites, including the Hoogsteen site, and not obey the simple rules required for genetics.
But the failure of the synthesis yielded a still more fundamental role for the repeating charge in a DNA molecule, one that suggested that repeating backbone charges were necessary for any biopolymer to support Darwinian evolution. Here, the argument is more subtle, and begins with the realization that replication alone is not sufficient for a genetic molecule to support Darwinian evolution. A Darwinian system must generate inexact replicates, descendants whose chemical structures are different from those of their parents. Further, these differences must then be replicable themselves. It does no good if the mutant has changed its biophysical properties so dramatically that the mutant genetic molecule precipitates, folds, or otherwise loses the ability to encode selectable information.
While self-replicating systems are well known in chemistry, those that generate inexact replicas with the inexactness itself being replicable are not [39]. As a rule, changing the structure of a molecule changes its physical behavior. Indeed, it is quite common in chemistry for small changes in molecular structure to lead to large changes in physical properties. This is certainly true in proteins, where a single amino acid replacement can cause the protein molecule to precipitate (the archetypal example of this is sickle cell hemoglobin). This means that inexact replicates need not retain the general physicochemical properties of their ancestors, in particular, properties that are essential for replication.
This thought, again arising through the analysis of a failed synthesis, led us to realize that a repeating backbone charge might be universal for all genetic molecules that work in water, on Earth, Mars, and Titan, but also for Vulcans, Klingons, and the Borg living in the Delta Quadrant of the Milky Way Galaxy, and even for Jedi living in a galaxy far, far away. The polyanionic backbone dominates the physical properties of DNA. Replacing one nucleobase in the sequence of a DNA molecule by another therefore has only a second order impact on the physical behavior of the molecule. This allows nucleobases to be replaced during Darwinian evolution without losing properties essential for replication.
This thought also puts in context the statement that DNA and RNA are “the same, except” for a replacement of an –H group by an –OH group on each of its biobricks. Such a change would have a major impact on the behavior of almost any other molecular system. It does not for DNA and RNA because their repeating backbone charges so dominate their overall behavior of these molecules that the changes expected through replacement of an –H by an –OH in each biobrick are swamped by the repeating charge. To this comment should be added the remark: “But only barely so”. DNA and RNA still have many differences in their physical properties that can be attributed to the replacement of an –H by an –OH in each biobricks.
We can bring our engineering synthetic biologist friends on board at this point. In the language of modern engineering synthetic biology, the repeating charge in the DNA backbone allows nucleotides to behave largely as interchangeable parts. It allows the whole to be the sum of its parts. It allows engineers, even those totally unfamiliar with Structure Theory, to design DNA molecules that pair with other DNA molecules according to simple rules. Because of this repeating backbone charge, and only because of this repeating backbone charge, is it possible to make “tiles” or biobricks from DNA, for example.
And only because of this repeating backbone charge can DNA and RNA support Darwinian evolution. The sequence ATCCGTTA behaves in most respects the same way as the sequence GCATGACA, even though these have very different molecular structures. This is because in both cases, the molecules are polyanions. These differences hold the genetic information. Were it otherwise, we could not mutate ATCCGTTA to give GCATGACA, even if GCATGACA better allowed us to survive, get married, and have children.
For this reason, the second-generation model for DNA proposed that a repeating charge should be a universal structure feature of any genetic molecule that supports Darwinian evolution in water, regardless of where it is found on Earth [34]. Polycationic backbones are also predicted to be satisfactory under what has become called the “polyelectrolyte theory of the gene” [34]. Thus, if NASA missions do detect life in water on other planets, their genetics are likely to be based on polyanionic or polycationic backbones, even if their nucleobases and sugars differ from those found on Earth. This structural feature can be easily detected by simple instruments, some of which might eventually fly to Mars or Titan.
Again, it is hard to believe that these insights would have emerged without synthetic biology. After all, first generation Watson-Crick theory had been in textbooks for three decades without recognizing the fundamental role of the repeating charge to the ability of DNA strands to bind their complements and support Darwinian evolution. Lacking that recognition, venture capitalists and other investors had bet billions of dollars on one particular antisense strategy, the one that required that molecular recognition remain in DNA analogs after the repeating charge was removed. Had they had the polyelectrolyte theory of the gene at their disposal, they would not have lost so much money. Synthesis drives discovery and paradigm changes in ways that analysis cannot.
3 Building genetics from the universal atom up
One not trained in synthesis might not have expected that failure could be so rewarding. We did not generate flexible DNA. We did not generate antisense drugs. But we did show that our theory of the molecular structures behind the most fundamental of biological processes, replication and evolution, was inadequate. This led to a better theory.
3.1 Could base pairing behind Darwinian evolution be so simple?
But what about the nucleobases, which had long been understood to be critical to the biological properties of DNA? And what about the simple rules that were proposed by Watson and Crick to account for genetics and Darwinian evolution: big pairs with small and hydrogen bond donors pair with hydrogen bond acceptors?
Could things be so simple? Again, if they were, then the synthetic biology paradigm (in the chemists’ sense of the contranym) laid before us a grand challenge. On paper, if we shuffled the red hydrogen bond donor and blue hydrogen bond acceptor groups in the A:T and G:C pairs (Fig. 7), treating these as interchangeable parts, we could write down eight new nucleobases that fit together to give four new base pairs having the same geometry as the A:T and G:C pairs (Fig. 8). Photocopy the page from this book, cut out the non-standard base pairs shown in Fig. 8, and fit them together yourself as a modern James Watson. As with the four standard nucleobases examined by Watson and Crick, the new nucleobases were predicted to pair with size complementarity (large with small) and hydrogen bond complementarity (hydrogen bond donors with acceptors), if the theory behind the pairing was so simple.

The two standard Watson-Crick pairs, idealized by replacing natural adenine (which lacks the bottom NH2 group) with amino adenine.

Shuffling hydrogen bond donor and acceptor groups in the standard nucleobase pairs generated eight additional heterocycles that, according to simple theory, should form four new, mutually independent, base pairs. This is called an “artificially expanded genetic information system” (AEGIS). Could molecular behavior at the center of genetics and Darwinian Evolution be so simple? Synthesis was used to decide.
As before, it was not enough to model the design on paper. Or even by computer. We needed to use synthetic technology from organic chemistry to create these new forms of matter, put them into DNA molecules, and see whether they worked as part of an artificially expanded genetic information system (AEGIS).
I will not leave you in suspense. Using the synthetic technology developed and enjoyed by chemists over the previous century, we were able to synthesize all of the synthetic components of our new artificial genetic alphabet. We were then able to put these synthetic nucleotides into synthetic DNA and RNA strands, and do all of the characterization of these that chemists do.
Once the synthetic task was complete, we observed that our artificial synthetic genetic system worked, and worked well. Artificial synthetic DNA sequences containing the eight new synthetic nucleotides formed double helices with their complementary synthetic DNA sequences. Complementation followed simple rules; just as A pairs with T and G pairs with C, P pairs with Z, V pairs with J, X pairs with K, and isoG pairs with isoC. The synthetic large nucleotides paired only with the correct synthetic small nucleotide. Our artificial synthetic DNA worked as well as natural DNA, at least in its ability to pair following simple rules.
An interesting irony is embedded in these results from synthetic biology. The base pairs were at the center of the Watson-Crick first generation model for duplex structures; the phosphates and the sugars were not. Once synthetic biologists got their hands on this molecule, it was found that the base pairs were the easiest to change.
3.2 Synthetic genetic systems to make synthetic protein systems
Synthetic genetic systems can pair. But can they meet advanced challenges? Again, when the synthesis successfully meets the grand challenge originally laid out, discipline requires us to assume that we have not been sufficiently ambitious in the selection of the challenge. Accordingly, we must next make the challenge more difficult. For example, could the extra nucleotides be used in a natural translation system to increase the number of amino acids that could be incorporated into proteins by encoded ribosome-based protein synthesis?
Meeting this challenge required more synthesis, of transfer RNA molecules carrying AEGIS nucleotides in the anticodon loop charged with a non-standard amino acid and of messenger RNA that contained the complementary non-standard AEGIS nucleotides. Again, the synthesis based on simple molecules of ribosome-catalyzed protein synthesis was adequate as a guide; the challenge was met and worked [40] (Fig. 9). Expanding the number of biobricks in synthetic DNA could also expand the number of biobricks in encoded proteins. Further, the fact that the theory was adequate to meet this challenge constitutes support for the theory, under an inversion of the Feynman dictum (“What I can make, I understand”).
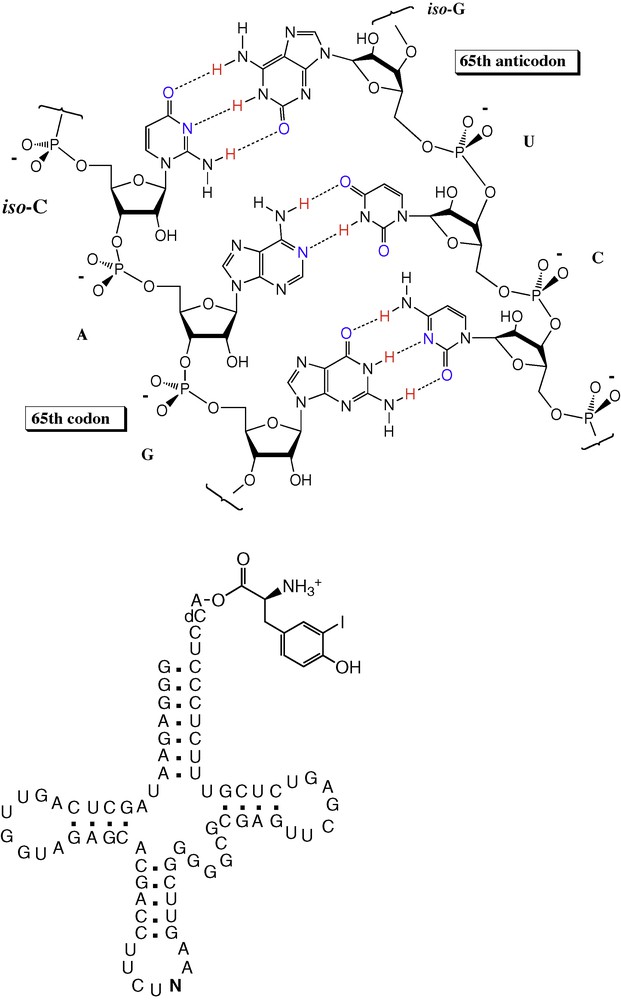
Putting a synthetic base into a messenger RNA, and providing a transfer RNA having the complementary non-standard base in the anticodon loop (the “N”) allowed the incorporation of a 21st amino acid (here, iodotyrosine) into a protein.
It should be noted that through this success with unnatural biology, something was learned about natural biology. As a control in one of the experiments with a messenger RNA molecule carrying an AEGIS base, we left out the charge transfer RNA having a non-standard nucleobase in its anticodon loop. We expected the synthesis of protein to stop at this point. Surprisingly, it did not. Instead, the ribosome paused, then skipped over the non-standard codon via a frame shift, and continued translation. This does not happen with standard stop codons built from standard nucleobases. This contrast in the behavior between the synthetic and natural systems shed new light on the way in which natural genetic systems terminate protein synthesis [34].
3.3 Synthetic genetics supports human health care
Pursuit of these “put-a-man-on-the-moon” challenges had taught us something. Base pairing is as simple from a molecular perspective as the first generation theory proposed in 1953 by Watson and Crick implied. Simple theories used by Watson and Crick, together with the new polyelectrolyte theory of the gene, were sufficient to empower the design of a new genetic system that works as well as natural DNA. Thus, these simple theories delivered an understanding of the molecular behavior of natural DNA. They also provided the language sufficient to explain genetics. As genetics is a big part of Darwinian evolution, synthesis made a big contribution to our understanding of life, at least under our definition-theory of life as a chemical system capable of Darwinian evolution.
Of course, these successes required us to again set the bar higher. Perhaps the best demonstration of our better understanding of DNA is to use it to create new technology; again mutating the Feynman dictum, we might suggest: “If we understand it, we can do something with it”.
Therefore, we set out to apply our synthetic genetic system in the clinic to support the care of human patients. The details are beyond the scope of this talk, but the general strategy is not. It might be worth a few words to explain how synthetic biology of this type has practical value.
Very often, diseases are caused by unwanted DNA. AIDS, for example, is caused by the human immunodeficiency virus (HIV), which delivers its own nucleic acid (RNA) into your body. A strep throat comes from unwanted bacteria carrying their unwanted DNA. Cancer comes from DNA from your own body that has mutated to give an unwanted sequence.
For such diseases, diagnosis involves detecting the unwanted DNA in a sample taken from a patient. But how can we find the unwanted DNA from the virus or the bacterium in that sample? After all, the unwanted DNA is present as just a few molecules in the sample; those few molecules are swamped by a background filled with considerable amounts of wanted DNA, the DNA from you the patient.
Accordingly, a general approach to detecting unwanted DNA involves two steps: (i) we must bind something to the unwanted DNA to form a bound complex; then (ii) we must move the bound complex to enrich and concentrate it at a spot where it can be detected.
Designing something to bind to unwanted DNA is easy if we know the sequence of the unwanted DNA. Following Watson-Crick pairing rules, we simply design a DNA strand that places a complementary A in a position where it can pair to each T in the unwanted DNA, a complementary T to pair with each A, a complementary C to pair with each G, and a complementary G to pair with each C. To illustrate with a trivial example, if the virus DNA sequence is TAAGCTTC, the DNA sequence GAAGCTTA will bind to it, and bind to it selectively. If you have difficulty seeing this, remember that one of the sequences binds to the other in reverse order. This is, of course, the same idea as was pursued in the antisense industry.
To concentrate the bound complex at a spot in a detection architecture, it would be nice to do the same trick: place a DNA molecule with specific capture sequence at that spot and then place the complementary tag on the bound complex containing the unwanted DNA. The tag would drag the unwanted DNA to that spot, where it could be detected.
We make the tags from A, T, G, and C, and drag the tags to the detection spot using A:T and G:C pairing. Solving the “how to move DNA around” problem in this way encounters a problem in any real assay, however. This problem arises because biological samples that are actually examined in the clinic (your blood, for example) contain lots of DNA containing lots of A, T, G, and C. While the tag would be designed to have a different sequence than the sequence of the wanted DNA that is in your blood, it is difficult with A, T, G, and C to make a tag that is very different. For example, your DNA has just about every sequence 15 nucleotides long built from A, T, G, and C. These sequences will interfere with capture and concentration of unwanted DNA at a spot when the tag is built from the natural nucleotides A, T, G and C.
This problem can be solved by incorporating the extra synthetic nucleobases into the capture and concentration tags. This is exactly what was done by Mickey Urdea and Thomas Horn as they were developing at Chiron a system to detect human immunodeficiency virus in the blood of AIDS patients. They used two of our synthetic nucleobases from the synthetic genetic alphabet (isoC and isoG, Fig. 8) to move bound unwanted DNA to a place where it could be detected, exploiting pairing between two complementary components of the synthetic genetic system that do not pair with the natural A, T, G, and C. This left A, T, G, and C available to bind to the unwanted DNA directly.
Because neither isoG nor isoC is found in the wanted DNA from the human patient, the large amount of background DNA cannot interfere with the capture of the unwanted DNA attached to the probe. This reduces the level of “noise” in the system. As a result using our synthetic genetic system, a diagnostic tool can detect as few as a dozen molecules of unwanted DNA in a sample of patient blood even though that blood is full of wanted DNA from the patient. Together, this assay measured the level of the RNA from human immunodeficiency virus (HIV, the causative agent of AIDS) in the blood of a patient, a measurement that allows the physician to adjust the treatment of the patient on a personal level to respond to the amount of virus that the patient has [41].
A similar diagnostic tool uses our synthetic genetic system to personalize the care of patients infected with hepatitis B and hepatitis C viruses. Still other applications of our synthetic genetic system are used in the analysis of cystic fibrosis, respiratory infections, influenza and cancer. Today, our synthetic genetic systems help each year to personalize the care of 400,000 patients infected with HIV and hepatitis viruses. With the support of the National Human Genome Research Institute, we are developing tools that will allow synthetic genetics to sequence the genomes of patients rapidly and inexpensively. These tools will ultimately allow your physician to determine rapidly and inexpensively the genetic component of the malady that afflicts you.
The fact that synthetic genetic systems empower commercial activity as well academic research makes us still more confident of the theory that underlay the synthetic effort in the first place. The kind of confidence comes from making something entirely new that not only works in the laboratory but also helps sick people; it is difficult to imagine a stronger way to obtain this confidence. In addition to driving discovery and paradigm change in ways that analysis cannot, synthetic biology allows us to generate multiple experimental approaches to decide whether our underlying view of reality is flawed.
3.4 The next challenge. Can artificial synthetic genetic systems support Darwinian evolution?
But why stop here? The next challenge in assembling a synthetic biology requires us to have our synthetic genetic system support Darwinian evolution. For this, we needed technology to copy synthetic DNA. Of course, copying alone would not be sufficient. The copies must occasionally be imperfect and the imperfections must themselves be copyable.
To copy our synthetic genetic system in pursuit of this goal, we turned to enzymes called DNA polymerases. Polymerases copy standard DNA strands by synthesizing new strands that pair A with T, T with A, G with C, and C with G [42]. The polymerases can then copy the copies, and then the copies of the copies. If done many times, this process is called the polymerase chain reaction, or PCR. PCR was developed by Kary Mullis, who was also awarded a Nobel prize.
As we attempted to meet this grand challenge, we immediately encountered an unscripted problem. Natural polymerases have evolved for billions of years to accept natural genetic systems, not synthetic genetic systems. As we tried to use natural polymerases to copy our synthetic DNA, we found that our synthetic DNA differed from natural DNA too much. Natural polymerases therefore rejected our synthetic DNA as “foreign”.
Fortunately, synthetic methods available from classical synthetic biology allow us to replace amino acids in the polymerases to get mutant polymerases. Several of these synthetic polymerases were found to accept our synthetic DNA. Michael Sismour and Zunyi Yang, working in my group, found combinations of polymerases (natural and synthetic) and synthetic genetic alphabets that worked together.
And so we went back to the laboratory to see if DNA molecules built from synthetic nucleotides could be copied, whether the copies could be copied, and whether the copies of the copies could be copied. We also asked whether the polymerases would occasionally make mistakes (mutations), and whether those mistakes could themselves be copied.
To meet this challenge, we did accommodate a bit the preferences of DNA polymerases. These have evolved for billions of years on Earth to accept nucleobases that present electron density (the green lobes in Fig. 10) to the minor groove (down, in Fig. 10) of the double helix. Many of our synthetic nucleobases to not do this, but two do: Z and P (Fig. 10). These form a P:Z base pair that actually contributes to duplex stability more than the A:T and G:C pairs [43].
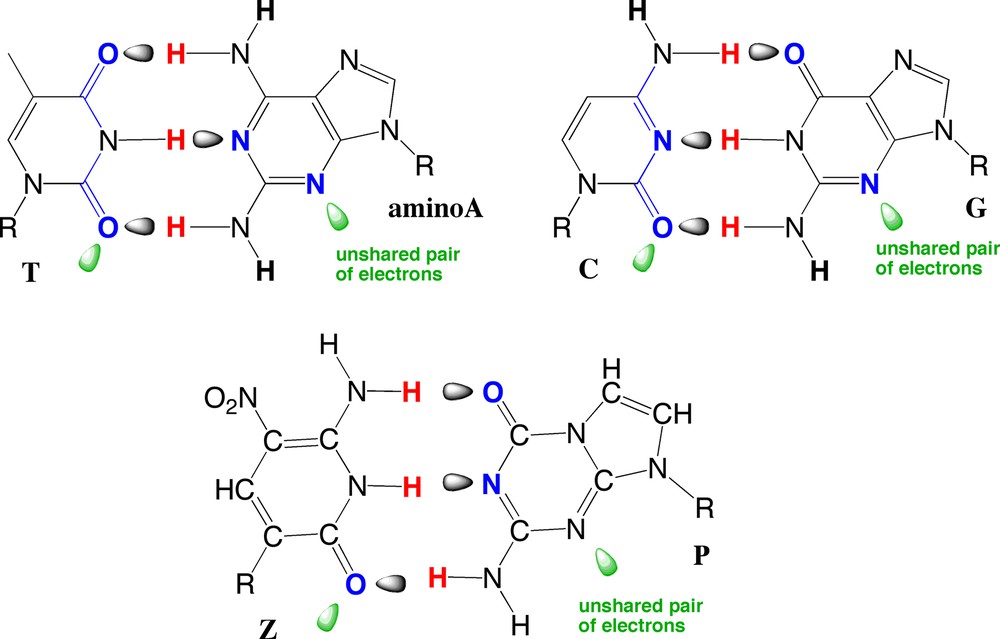
The Z and P pairs that have been incorporated into six-letter PCR, with mechanistic studies that show that this six-letter synthetic genetic system can support Darwinian evolution. Key to meeting this challenge was to make a small accommodation to the desire of natural DNA polymerases to have nucleobases that present electron density (the green lobes) to the minor groove (down, in this structure) of the double helix. This is the case for T, A (shown here with an extra NH2 unit), C, and G (top). It is also the case with the Z and P synthetic nucleobases (bottom).
But would they work with natural enzymes? Again, we do not want to keep you in suspense. A six-letter synthetic genetic system built from four standard nucleotides and two synthetic nucleotides can be repeatedly copied (Fig. 11).
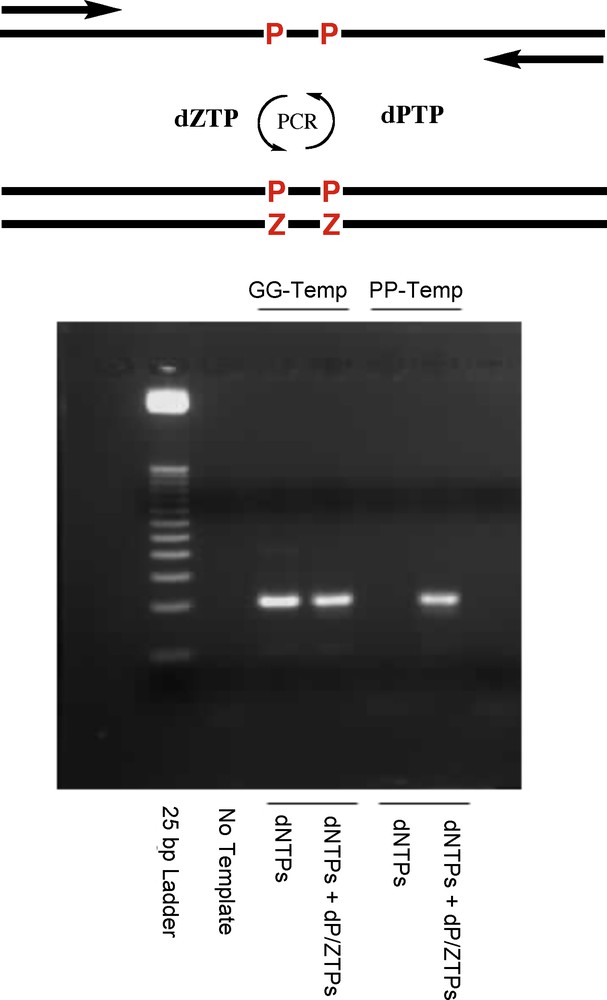
The polymerase chain reaction with a six-letter genetic alphabet, incorporating P and Z in addition to A, T, G, and C.
Setting the bar higher, could they support Darwinian evolution? Here, careful experiments were done to determine whether they could participate in mutation processes. This work showed that Z and P could indeed mutate to C and G and, more surprisingly, that C and G could mutate to give Z and P. The details of the mutation process were studied. Sometimes Z is incorporated opposite G instead of C. Sometimes C is incorporated opposite P instead of Z. Sometimes P is incorporated opposite C instead of G. Sometimes G is incorporated opposite Z instead of P. This low level of mutation is just a few percent per copy. But once mutations are introduced into the children DNA, they themselves can be copied and therefore propagated to the next generation. Thus, the synthetic genetic system built from G, A, C, T, Z and P (GACTZP) is capable of supporting Darwinian evolution.
3.5 Is this synthetic life?
A GACTZP synthetic six-letter genetic system that includes the “biobricks” G, A, C, T, Z and P is clearly not homologous to the genetic system that we find naturally on Earth. It does, of course, share many structural features with natural genetic systems. Some of these we believe to be universal based on theories like the polyelectrolyte theory of the gene. The repeating backbone phosphates are not, according to that theory, dispensable.
But it can support Darwinian evolution. Is this artificial synthetic life? Our theory-definition holds that life is a self-sustaining chemical system capable of Darwinian evolution. The artificial genetic system that we have synthesized is certainly a chemical system capable of Darwinian evolution. It is not self-sustaining, however. For each round of evolution, a graduate student must add something by way of food; the system cannot go out to have lunch on its own.
Therefore, while our synthetic genetic system demonstrates that simple theory can empower/explain the molecular side of evolution, we are not yet at the point where we can use our synthetic genetic as a “second example” of life. We are not ready to use our system to see whether it can spontaneously generate traits that we recognize from natural biology.
For this, we return to the need for bucks. Not surprisingly (and not inappropriately), funding is easier to find to research tools that help manage the medical care of patients infected with HIV and other viruses than to take synthetic biology the next step. Because of the relevance of this work to these applications work is proceeding. Further, the Defense Threat Reduction Agency is encouraging us to develop further the second-generation model of DNA.
Accordingly, we are attempting to meet the “put-a-man-on-the-moon” goal of obtaining a synthetic genetic system that can sustain to a greater degree its own access to Darwinian evolution.
Even should this be done, however, the community would not be unanimous in its view that a synthetic biology had been created. Various theories of biology constructively held by many in the community add criteria for a definition-theory of life. For example, those who subscribe to the Cell Theory of life will no doubt wait until the synthetic chemical system capable of Darwinian evolution is encapsulated in a cell. Those who subscribe to a metabolism theory of life might wait until the artificial synthetic genetic system also encodes enzymes that catalyze the transformation of organic compounds. Even those who subscribe to a Darwinian theory of life might insist that before a synthetic biology is announced, the artificial system must evolve to a natural change in environment, not to one engineered in the laboratory.
Where one draws the line is, again, a matter of culture. Further, we expect that as these goals are pursued, the bar will be raised; again, if success is achieved, the discipline of a synthetic biologist requires the bar to be raised.
Again, this is not relevant to the value of the pursuit. The purpose of the synthetic effort is to force ourselves across uncharted territory where we must address unscripted questions. As we attempted to design a synthetic genetic system or a synthetic protein catalyst, we learned about genetics and catalysis in general, as well as the strengths and inadequacies of our theories that purported to understand these. In future pursuits of synthetic cells, metabolisms, and adaptation in a synthetic biological system, we cannot help but learn more about cells, metabolism, and adaptation in general, including these processes found naturally in life around us today on Earth. Illustrated by the four-wedge diagram in Fig. 2, learning from synthesis will complement learning obtained from paleogenetics, exploration, and laboratory experiments attempting to understand the origin of life.
4 Does synthetic biology carry hazards?
Provocative titles like “synthetic biology” and “artificial life” suggests a potential for hazard. They also conjure up images of Frankenstein. Is there any hazard associated with synthetic biology? If so, can we assess its magnitude?
As noted in the introduction, much of what is called “synthetic biology” today is congruent with the activities supported by the recombinant DNA technology that has been around for the 35 years since Waclaw Szybalski coined the term. There is no conceptual difference between how bacteria are constructed today to express genes from other places and how they were constructed in 1980; a straight line connects the synthetic biology of Szybalski to the current efforts of Venter, Smith and their colleagues.
The hazards of this type of synthetic biology were discussed at a famous 1975 conference at the Asilomar conference site in Monterey, California. We now have a quarter century of experience with the processes used to mitigate any hazards that might exist from this type of synthetic biology. Placing a new name on an old research paradigm does not create a new hazard; much of the concern over the hazards of today's efforts of this type reflect simply their greater chance of success because of improved technology.
Those seeking to create artificial chemical systems to support Darwinian processes are, however, creating something new. We must consider the possibility that these artificial systems might escape from the laboratory. Does this possibility create a hazard?
Some general biological principles are relevant to assessing the potential for such hazards. For example, the more an artificial living system differs (at a chemical level) from a natural biological system, the less likely it is to survive outside of the laboratory. A living organism survives when it has access to the resources that it needs, and is more fit than competing organisms in recovering these resources from the environment where it lives. Thus, a completely synthetic life form having unnatural nucleotides in its DNA would have difficulty surviving if it were to escape from the laboratory. What would it eat? Where would it get its synthetic nucleotides?
Such principles also apply to less exotic examples of engineered life. Thirty years of experience with genetically altered organisms since Asilomar have shown that engineered organisms are less fit than their natural counterparts to survive outside of the laboratory. If they survive at all, they do so either under the nurturing of an attentive human or by ejecting their engineered features.
Thus, the most hazardous type of bioengineering is the type that is not engineering at all, but instead reproduces a known virulent agent in its exact form. The recent synthesis of smallpox virus or the 1918 influenza virus are perhaps the best examples of risky synthetic biology.
Further, we might consider the motivation of one actually wanting to do damage? Would one generate a genetically engineered Escherichia coli? Or place fuel and fertilizer in a rented truck and detonate it outside of the Federal Building in Oklahoma City? We know the answer to this question for one individual. We do not know it for all individuals. In most situations, however, it seems easier to do harm in non-biotechnological ways than by engineering biohazards.
Any evaluation of hazard must be juxtaposed against the potential benefits that come from the understanding developed by synthetic biology. History provides a partial guide. In 1975, the City of Cambridge banned the classical form of synthetic biology within its six square miles to manage what was perceived as a danger. In retrospect, it is clear that had the ban been worldwide, the result would have been more than harmful. In the same decade that Cambridge banned recombinant DNA research, an ill-defined syndrome noted in patients having “acquired immune deficiency” was emerging around the planet as a major health problem. This syndrome came to be known as AIDS, and it was eventually learned that AIDS was caused by the human immunodeficiency virus (HIV).
Without the technology that the City of Cambridge banned, we would have been hard pressed to learn what HIV was, let alone have compounds today that manage it. Today, classical synthetic biology and recombinant DNA technology allows us to manage new threats as they emerge, including SARS, bird influenza, and other infectious diseases. Indeed, it is these technologies that distinguish our ability to manage such threats today from how we would have managed them a century ago.
With these thoughts in mind, a Venn diagram can be proposed to assess risk in different types of synthetic biology. Activities within the red circle use standard terran biochemistry, more or less what Nature has developed on Earth over the past four billion years. Activities outside that circle concern activities with different biochemistry (Fig. 12).
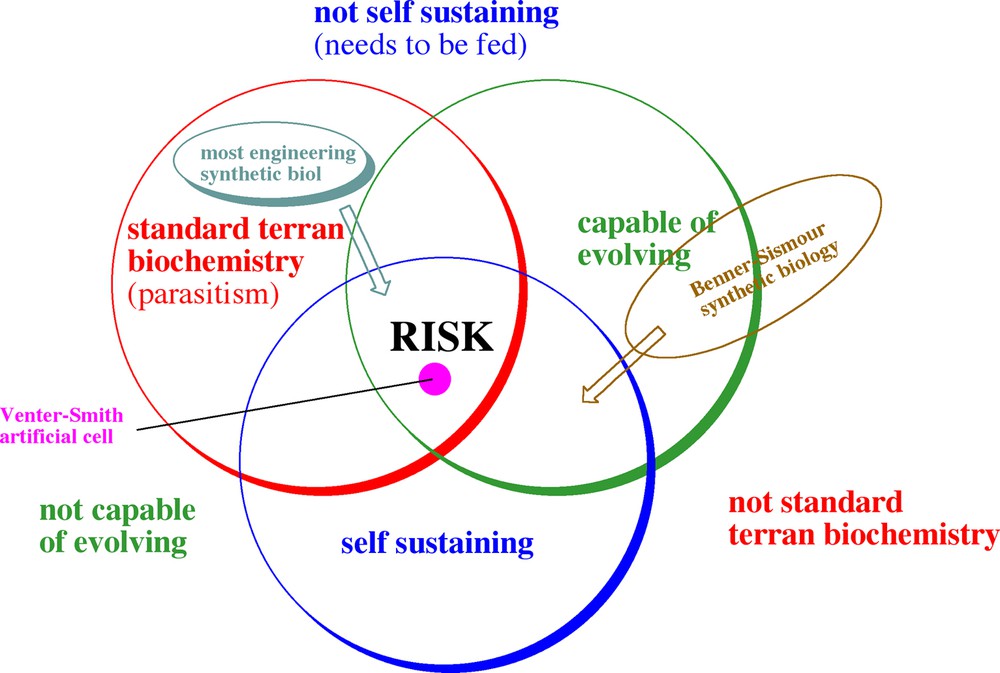
A Venn diagram illustrating the hazards of synthetic biology. The green circle contains systems able to evolve. Those outside the circle cannot, and present no more hazard than a toxic chemical. The blue circle contains systems that are self-sustaining. Those inside the circle “live” without continuing human intervention; those outside require continuous feeding, and are no more hazardous than a pathogen that dies when released from a laboratory. Systems within the red circle use standard terran molecular biology; those outside do not. The greatest chance for hazard comes from a system that is self-sustaining, uses standard biochemistry, and is capable of evolving, the intersection between the three circles.
The green circle contains systems that are capable of evolving. Those outside the circle cannot, and present no more hazard than a toxic chemical; regardless of its hazard, it is what it is, and cannot get any worse.
The blue circle contains systems that are self-sustaining. They “live” without continuing human intervention. Those outside the blue circle require continuous feeding. Thus, these represent no more of a hazard than a pathogen that will die once released from the laboratory.
The greatest chance for hazard comes from a system that is self-sustaining, uses standard biochemistry, and is capable of evolving. This is, of course, the goal of the Venter-Smith artificial cell, which presents the same hazards as presented by natural non-pathogenic organisms: it might evolve into an organism that feeds on us. Those hazards, although not absent, are not large compared to those presented by the many natural non-pathogens that co-inhabit Earth with us.
5 Conclusions
Pursuit of the grandest challenge in contemporary synthetic biology, creating artificial life of our own, has already yielded fruits. Alien life with six letters in its genetic alphabet and more than 20 amino acids in its protein alphabet is possible. A repeating charge may be universal in the backbone of genetic biopolymers; a repeating dipole may be universal in the backbone of catalytic biopolymers. We have synthesized in the laboratory artificial chemical systems capable of Darwinian evolution.
This makes the next grand challenge still more ambitious. We would like a self-sustaining artificial chemical system capable of Darwinian evolution. To get it, the reliance of the current synthetic Darwinian systems on natural biology must be reduced. We have encountered unexpected problems as we attempt to do so. Some in the community are confident that with a little more effort, we can surmount these problems; others of us are not so sure.
Achieving in the laboratory an artificial biology would expand our knowledge of life as a universal more than anything else short of actually encountering alien life. Still better, it is more likely that synthetic biology will do this sooner than exploration will. If we had a simple form of designed life in our hands, we could ask key questions. How does it evolve? How does it create complexity? How does it manage the limitations of organic molecules related to Darwinian processes? And if we fail after we give the effort our best shot, it will directly challenge our simple (and possibly simplistic) definition-theory of life as being nothing more than a chemical system capable of Darwinian evolution.
Conflict of interest statement
Various authors are inventors on patents covering certain of these technologies.
Acknowledgements
We are indebted to the NHGRI (R01HG004831, R01HG004647) and the NIGMS (R01GM086617) for support of the applied parts of this work, the NASA Astrobiology program for support of aspects of work relating to discovery of alien life (NNX08AO23G), and the Defense Threat Reduction Agency (HDTRA1-08-1-0052) in its basic research program. We are especially indebted to the encouragement of DTRA to develop the basic theory of DNA.


