1 Introduction
The aqueous solution chemistry of Pt(II) complexes has been extensively studied during recent decades, owing to the anticancerous activity of cisplatin, cis-diamminedichloroplatinum(II), cis-Pt(NH3)2Cl2 and related compounds [1–5]. The antitumour activity of cisplatin is ascribed to interactions between the complex and DNA [2–4]. There are many other potential biomolecules that can also react with these Pt(II) complexes, such as small molecules, proteins and enzymes. In fact, already in the blood, where the Pt drug is administrated by injection or infusion, several molecules are available for thermodynamic competition [3,4]. Although, cisplatin is one of the most effective antitumor agents, there are still difficulties related to its use, because of numerous side effects and its toxicity. Replacement of chloro ligands by carboxylate groups in carboplatin [6], leading to cis-diamine(1,1-cyclobutanedicarboxylate)-platinum(II) reduces the toxicity of the complex and increases its solubility in water. Some investigators have suggested that carboplatin is merely a pro-drug for cisplatin [7], whereas others have postulated a ring opening reaction [8] of carboplatin followed by the reaction with DNA. Therefore the thermodynamics of the reactions for the Pt(II) complexes is of great interest. In order to avoid the inert substitution behavior of Pt(II) complexes, and on the basis of the remarkable analogy between the coordination chemistry of Pt(II) and Pd(II) complexes, a series of labile Pd(II) complexes have proved useful as models for the Pt(II) complexes.
Work in our laboratories focused on the studies of metal complexes of biological significance [9–15]. Pd(II) complexes with bidentate amine forming a five-membered chelate ring were extensively investigated as a model of the antitumor cis-diaminePt(II) complex. It seemed of interest to study the behavior of palladium(II) complexes with 1,3-diamino-2-hydroxopropane, involving a six-membered chelate ring. This may be the first step in the development of a new generation of antitumor agents. Also, the increase in the chelate ring size will increase the bite angle, which have an effect of increasing the steric interaction between the guanines in the cis-Pt(diamine)G2 adduct, thereby slowing down the rotation of the guanines about the Pt-N7 bonds [16,17]. Such restriction may stabilize the DNA adduct. This is in line with the finding that cis-Pt(1,4-DACH)Cl2, (1,4-DACH = 1,4-diaminocyclohexane) where a seven-membered chelate ring is formed, is more active than cisplatin and oxaliplatin in several in vivo and in vitro tests [18]. Also, the OH group may undergo hydrogen bonding with DNA. Such effects may favor the interaction with DNA, which is the main target for the antitumor agent. For these reasons, it seems therefore of considerable interest to perform a systematic study of the complex formation equilibria between [Pd(DHP)(H2O)2]2+ and amino acids, peptides, dicarboxylic acids or DNA constituents.
2 Experimental
2.1 Materials
PdCl2 and 1,3-diamino-2-hydroxopropane (DHP) were obtained from Aldrich Chem. Co. The dicarboxylic acids, amino acids, peptides and DNA constituents investigated are: 1,1-cyclobutanedicarboxylic acid, oxalic acid, malonic acid succinic acid, adipic acid, glycine, alanine, proline, methionine, ornithine.HCl, histamine.2HCl, histidine.HCl, serine, leucine, cysteine.HCl, glycinamide, glycylglycine, glutamine, aspargine, uracil, uridine, thymine, thymidine and guanosine 5’-monophosphate disodium salt. These materials were provided by Sigma Chemical company and used without further purification. Carbonate-free NaOH (titrant) was prepared and standardized against potassium hydrogen phthalate solution daily. All solutions were prepared in deionized H2O.
2.2 Synthesis
Pd(DHP)Cl2 was prepared by dissolving K2PdCl4 (2.82 mmol) in 10 ml water with stirring. The clear solution of [PdCl4]2− was filtered and 1,3-diamino-2-hydroxopropane (2.82 mmol), dissolved in 10 ml H2O was added drop wise to the stirred solution. The pH was adjusted to 2–3 by the addition of HCl and/or NaOH. A yellowish -brown precipitate of Pd(DHP)Cl2 was formed and stirred for a further 30 minutes at 50 °C. After filtering off the precipitate, it was thoroughly washed with H2O, ethanol and diethyl ether. A yellow powder was obtained. Anal. Calcd. for C3H10N2OPdCl2 (267.3):C, 13.6; H, 3.7; N, 10.5. Found: C, 13.5; H, 4.0; N, 10.3%.
Pd(DHP)(CBDCA) was synthesised by stirring of Pd(DHP)Cl2 complex (0.267 g, 1.0 mmol) with Ag NO3 (0.168 g, 1.98 mmol) in 10 ml H2O overnight in the dark. After filtering off the white precipitate (AgCl), a solution of H2CBDCA (0.144 g, 1.0 mmol) dissolved in 20 ml H2O was added to the yellow filtrate. The pH value was adjusted to 5.0 with NaOH and the solution mixture was stirred for 2 h. At 60 °C. After storing at 4 °C for 5 days, pale yellow crystals were obtained, isolated and washed with H2O, ethanol and finally with diethyl ether. (yield 65%), Anal. Calcd. For C9H16N2O5Pd (338.4): Calcd. C 31.9, H 4.7, N 8.3. Found C 31.6, H 4.9, N 8.1%.
The ligands in the form of hydrochlorides were converted to the corresponding hydronitrate in the same way as described above
2.3 Apparatus
Potentiometric measurements were conducted using a Metrohm 686 titroprocessor equipped with a 665 Dosimat (Switzerland-Herisau). A thermostated glass-cell was used equipped with a magnetic stirring system, a Metrohm glass-calomel combined electrode, a thermometric probe and a microburete delivery tube. The titroprocessor and electrode were calibrated daily with standard buffer solutions prepared according to NBS specifications at 25.0 ± 0.1 °C [19] and I = 0.1 mol-dm−3, potassium hydrogen phthalate (pH 4.008) and a mixture of KH2PO4 and Na2HPO4 (pH 6.865). Elemental microanalyses of the separated solid for C, H and N was performed in the Microanalytical Center, Cairo University. The analyses were performed twice to check the accuracy of the analyses data.
2.4 Procedure and measuring techniques
Determination of equilibrium binding constants in palladium(II) coordination complexes presents a challenge due to the lack of convenient spectroscopic handles. We found that it was possible to obtain the data by potentiometric methods. A typical experiment consisted of three titrations plus data analysis. First, a solution of free ligand in the protonated form (0.125 mM) was titrated to obtain proton dissociation constants. These constants are essential for data analysis. A solution of [Pd(DHP)(H2O)2)]2+ (0.125 mM) was titrated to obtain the acid-dissociation constants of coordinated water. A solution mixture of [Pd(DHP)(H2O)2)]2+ (0.125 mM) and the ligand in concentration ratios of 1:1 for dicarboxylic acids, amino acids and peptides, and for concentration ratio of 1:2 (metal:ligand) for DNA constituents was titrated. A 0.05 M solution of NaOH was used as a titrant. The titration solution mixtures had a volume of 40 ml. Temperature was maintained constant inside the cell at (25.0 ± 0.1) °C, by circulating thermostated water through the double-wall titration vessel and under a slow and constant stream of N2 over the test solutions. The pH meter readings were converted into hydrogen ion concentration by titrating a standard acid solution (0.05 mol-dm−3), the ionic strength of which was adjusted to 0.1 mol-dm−3, with standard base solution (0.05 mol-dm−3) at 25 °C. The pH is plotted against p[H]. The relationship pH − p[H] = 0.05 was observed. [OH−] value was calculated using a pKw value of 13.997 [20]. The ionic strength was adjusted to 0.1 mol-dm−3 by using NaNO3. The equilibrium constants evaluated from the titration data (summarized in Table 1) are defined by Eqs. (1) and (2), where M, L and H stand for the [Pd(DHP)(H2O)2]2+ ion, ligand and proton, respectively. Charges are omitted for simplicity.
| (1) |
| (2) |
Formation constant of MpLqHr species in aqueous solution at 25 ± 0.1 °C and I = 0.1 mol-dm−3(NaNO3).
| System | p | q | ra | log βb | Sc |
| Pd(DHP)(OH2)2 | 1 | 0 | −1 | −5.61 (0.01) | 1.1E-7 |
| 1 | 0 | −2 | −15.53 (0.03) | ||
| 2 | 0 | −2 | −7.50 (0.02) | ||
| 1,1-Cyclobutanedicarboxylic acid | 0 | 1 | 1 | 5.74 (0.01) | 3.6E-8 |
| 0 | 1 | 2 | 9.20 (0.01) | ||
| 1 | 1 | 0 | 6.55 (0.01) | 1.1E-7 | |
| Oxalic acid | 0 | 1 | 1 | 4.01 (0.01) | 1.6E-8 |
| 0 | 1 | 2 | 6.55 (0.01) | ||
| 1 | 1 | 0 | 7.93 (0.03) | 2.7E-7 | |
| Malonic acid | 0 | 1 | 1 | 5.57 (0.01) | 1.6E-8 |
| 0 | 1 | 2 | 8.75 (0.07) | ||
| 1 | 1 | 0 | 5.89 (0.01) | 4.6E-8 | |
| Succinic acid | 0 | 1 | 1 | 5.42 (0.01) | 2.8E-8 |
| 0 | 1 | 2 | 9.66 (0.01) | ||
| 1 | 1 | 0 | 4.35 (0.01) | 3.4E-8 | |
| 1 | 1 | 1 | 8.63 (0.01) | ||
| Adipic acid | 0 | 1 | 1 | 5.57 (0.01) | 3.6E-8 |
| 0 | 1 | 2 | 9.61 (0.01) | ||
| 1 | 1 | 0 | 4.23 (0.01) | 1.1E-8 | |
| 1 | 1 | 1 | 8.49 (0.01) | ||
| Glycine | 0 | 1 | 1 | 9.67 (0.01) | 8.3E-8 |
| 0 | 1 | 2 | 12.15 (0.03) | ||
| 1 | 1 | 0 | 10.41 (0.02) | 1.2E-8 | |
| Alanine | 0 | 1 | 1 | 9.76 (0.01) | 4.9E-8 |
| 0 | 1 | 2 | 12.13 (0.01) | ||
| 1 | 1 | 0 | 10.58 (0.01) | 5.5E-8 | |
| Proline | 0 | 1 | 1 | 10.67 (0.01) | 1.3E-8 |
| 0 | 1 | 2 | 12.69 (0.05) | ||
| 1 | 1 | 0 | 11.10 (0.01) | 9.0E-8 | |
| Methionine | 0 | 1 | 1 | 9.15 (0.01) | 5.4E-8 |
| 0 | 1 | 2 | 11.37 (0.02) | ||
| 1 | 1 | 0 | 8.78 (0.02) | 1.4E-7 | |
| 0 | 1 | 1 | 10.66 (0.01) | 1.0E-8 | |
| Ornithine | 0 | 1 | 2 | 19.51 (0.01) | |
| 0 | 1 | 3 | 21.45 (0.01) | ||
| 1 | 1 | 0 | 13.02 (0.02) | 2.1E-8 | |
| 1 | 1 | 1 | 20.07 (0.01) | ||
| Histamine | 0 | 1 | 1 | 9.96 (0.01) | 2.4E-8 |
| 0 | 1 | 2 | 16.10 (0.01) | ||
| 1 | 1 | 0 | 12.68 (0.04) | 3.9E-7 | |
| 1 | 1 | 1 | 16.35 (0.08) | ||
| Histidine | 0 | 1 | 1 | 9.60 (0.01) | 1.8E-7 |
| 0 | 1 | 2 | 15.92 (0.03) | ||
| 1 | 1 | 0 | 13.99 (0.03) | 9.3E-7 | |
| Serine | 0 | 1 | 1 | 9.19 (0.01) | 1.7E-8 |
| 0 | 1 | 2 | 11.62 (0.01) | ||
| 1 | 1 | 0 | 10.01 (0.01) | 2.8E-8 | |
| 1 | 1 | −1 | 1.40 (0.01) | ||
| Leucine | 0 | 1 | 1 | 9.78 (0.01) | 3.4E-8 |
| 0 | 1 | 2 | 12.25 (0.01) | ||
| 1 | 1 | 0 | 10.39 (0.01) | 3.2E-8 | |
| 0 | 1 | 1 | 10.36 (0.01) | 2.9E-7 | |
| Cysteine | 0 | 1 | 2 | 18.01 (0.03) | |
| 0 | 1 | 3 | 20.62 (0.07) | ||
| 1 | 1 | 0 | 15.62 (0.04) | 2.1E-7 | |
| 1 | 1 | 1 | 19.83 (0.06) | ||
| Glycinamide | 0 | 1 | 1 | 7.89 (0.01) | 2.9E-8 |
| 1 | 1 | 0 | 7.77 (0.08) | 5.3E-7 | |
| 1 | 1 | −1 | 4.44 (0.02) | ||
| Glycylglycine | 0 | 1 | 1 | 7.98 (0.01) | 4.0E-8 |
| 1 | 1 | 0 | 7.27 (0.03) | 4.2E-7 | |
| 1 | 1 | −1 | 1.86 (0.03) | ||
| Glutamine | 0 | 1 | 1 | 8.93 (0.02) | 1.0E-8 |
| 1 | 1 | 0 | 9.47 (0.01) | 3.3E-8 | |
| 1 | 1 | −1 | −0.15 (0.01) | ||
| Aspargine | 0 | 1 | 1 | 8.93 (0.02) | 5.8E-8 |
| 1 | 1 | 0 | 9.25 (0.01) | 1.5E-7 | |
| 1 | 1 | −1 | 1.89 (0.02) | ||
| Uracil | 0 | 1 | 1 | 8.83 (0.01) | 1.2E-8 |
| 1 | 1 | 0 | 8.12 (0.01) | 5.8E-9 | |
| 1 | 2 | 0 | 14.21 (0.02) | ||
| Uridine | 0 | 1 | 1 | 9.01 (0.01) | 6.0E-8 |
| 1 | 1 | 0 | 8.35 (0.01) | 3.5E-8 | |
| 1 | 2 | 0 | 13.75 (0.01) | ||
| Thymine | 0 | 1 | 1 | 9.30 (0.01) | 7.5E-8 |
| 1 | 1 | 0 | 8.20 (0.02) | 1.5E-8 | |
| 1 | 2 | 0 | 13.72 (0.04) | ||
| Thymidine | 0 | 1 | 1 | 9.30 (0.01) | 7.5E-8 |
| 1 | 1 | 0 | 8.33 (0.02) | 1.6E-8 | |
| 1 | 2 | 0 | 13.28 (0.02) | ||
| Guanosine-5’-monophosphate | 0 | 1 | 1 | 9.48 (0.02) | 4.3E-8 |
| 0 | 1 | 2 | 15.82 (0.01) | ||
| 1 | 1 | 0 | 7.61 (0.02) | 5.1E-8 | |
| 1 | 2 | 0 | 10.97 (0.04) | ||
| 1 | 1 | 1 | 14.85 (0.02) | ||
| 1 | 1 | 2 | 20.76 (0.02) |
a p, q and r are the stoichiometric coefficients corresponding to Pd(DHP)2+, (amino acid, dicarboxylic acid, peptide or DNA) and H+, respectively.
b Standard deviations are given in parentheses.
c Sum of square of residuals.
The calculations were obtained from ca. 100 data points in each titration using the computer program MINIQUAD-75 [21]. The stoichiometry and stability constants of the complexes formed were determined by trying various possible composition models. The model selected gave the best statistical fit and was chemically consistent with the titration data without giving any systematic drifts in the magnitudes of various residuals, as described elsewhere [21]. The fitted model was tested by comparing the experimental titration data points and the theoretical curve calculated from the values of the acid dissociation constant of the ligand and the formation constants of the corresponding complexes. The results are summarized in Table 1. The species distribution diagrams were obtained using the program SPECIES [22] under the experimental condition employed.
2.5 Spectrophotometric measurements
Spectrophotometric measurements of Pd(DHP)-glycylgycine complex were performed by recording the UV-Visible spectra of solutions (A−C), where (A) = 1 mM of Pd(DHP)(H2O)22+; (B) = 1 mM of PdDHP)(H2O)22+ + 1 mM of glycylglycine + 1 mM of NaOH and (C) = 1 mM of Pd(DHP)(H2O)22+ + 1 mM of glycylglycine + 2 mM of NaOH. Under these prevailing experimental conditions and after neutralization of the hydrogen ions released, associated with complex formation, it is supposed that the complexes have been completely formed. In each mixture the volume was brought to 10 ml by addition of deionized water and ionic strength is kept constant at 0.1 M NaNO3.
3 Results and discussion
3.1 Characterization of the solid complexes
The IR spectra of the Pd(DHP)Cl2 and Pd(DHP)(CBDCA) complexes show a sharp band at 3414 cm−1 for stretching vibration of OH group and distinct bands at 3050 − 3250 cm−1, which can assigned to stretching vibrations of NH2 group. The complexes exhibit bands for (NH2) bending at 1466 and 1570 cm−1 and bands for the stretching vibration corresponding to Pd-N at 424 and 436 cm−1 for the previous complexes respectively. Band at 459 cm−1 is assigned to the stretching vibration of Pd-O for Pd(DHP)(CBDCA) complex. The bands at 1704 and 1288 cm−1 are attributed to the stretching vibrations of C = O and C-O for carboxylic group in free cyclobutanedicarboxylic acid. Upon coordination, the C = O band is shifted to lower frequency, 1635 cm−1, while the C-O band was shifted to higher frequency, 1386 cm−1. This indicates the participation of carboxylate group of CBDCA in complex formation.
The 1H-NMR spectrum of Pd(DHP)(CBDCA) complex was recorded. The H(β) and H(γ) protons of cyclobutane group in the free H2CBDCA appear at 2.38 and 1.85 ppm, respectively, corresponding well to literature values for related systems [9]. Due to coordination to Pd(II), the H(β) signal is shifted downfield by ∼0.45 ppm in comparison to the free ligand. On the other hand, the H(γ) signal is not significantly affected (downfield by ∼0.1 ppm). This may be explained in the premise that H(γ) is considerably far from the center of coordination.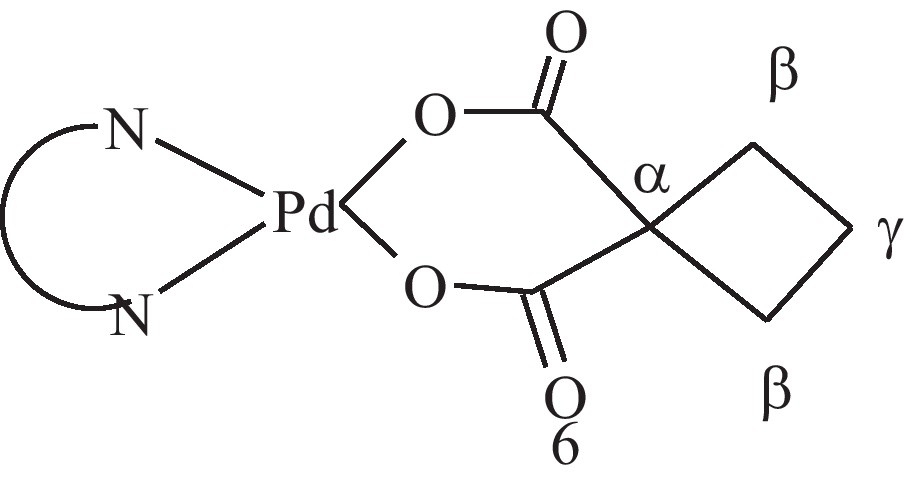
3.2 Acid-base equilibria
The acid dissociation constants of the ligands were determined under the experimental conditions of (25 ± 0.1) °C and a constant ionic strength of 0.1 mol-dm−3, which were also used to determine the stability constants of the Pd(II) complexes. The values obtained are consistent with data reported in the literature [23,24].
The acid-base equilibria of the [Pd(DHP)(H2O)2]2+ complex is characterized by fitting the potentiometric data to various models. The best-fit model was found to be consistent with the species 10-1 and 10-2 as given in Eq. (3) and the negative numbers refer to proton loss.
| (3a) |
| (3b) |
| (3c) |
The pKa1 and pKa2 values were found to be 5.61 and 9.92, respectively. The pKa1 value is higher than that for [Pd(AMBI)(H2O)2]2+ (pKa 4.7) [25] (AMBI = 2-aminomethylbenzimidazole). This shows that the first coordinated water molecule in [Pd(DHP)(H2O)2]2+ is less acidic than that of [Pd(AMBI)(H2O)2]2+. This is attributed to the π-acceptor properties of the aromatic moieties of AMBI, which leads to an increase in the electrophilicity of the Pd ion and consequently decreases the pKa of the coordinated water molecule. The equilibrium constant for the dimerization reaction (3) can be calculated by Eq. (4) and amounts to 3.72.
3.3 Complex-formation equilibria involving dicarboxylic acids
Analysis of pH titration data of Pd(DHP)-dicarbaoxylic acid system showed the formation of the 1:1 complex. The results (Table 1) show that the formation constant of the 1:1 complex involving formation of five and six membered chelate rings as in cyclobutanedicarboxylic acid, oxalic acid and malonic acid are higher than those involving seven-membered chelate ring, as in succinic acid and nine-membered ring as in adipic acid. This may be explained on the premise that the five-and six-membered rings are more favoured energetically than the seven- and nine-membered ring. It is interesting to note that CBDCA has a higher stability constant than malonic acid, although both of them form six-membered chelate ring. This may be due to the higher pKa values of the former than the latter dicarboxylic acid. Succinic acid and adipic acid form in addition to the 1:1 complex (110), the protonated form (111). The pKa of the protonated species (pKa = log β111 − log β110) is 4.28 for succinic acid and 4.26 for adipic acid. These values are close to the second acid dissociation constant of the corresponding acid. The distribution diagram of [Pd(DHP)]2+ − CBDCA complex was given in Fig. 1. The main species in the physiological pH range is the ring-closed form, 110, which reaches a maximum concentration of 98.5% at pH ca. 5.9.

Concentration distribution curves for Pd(DHP)CBDCA complexes.
3.4 Complex formation equilibria involving amino acids
Fitting of pH titration data for Pd(DHP)-amino acid equilibria indicated the formation of 1:1 complexes with high stability constant values. This indicates that amino acids bind through the amino and carboxylate groups. Serine has an extra binding center on the β-alcoholate group. This group was reported 26 to participate in transition metal ion complex formation reactions. The pKa value of the alcoholate group incorporated in the Pd(II) complex (log β110 − log β11-1) is 8.61. Therefore in physiological pH (7.4), the −OH group participates in bonding with Pd(DHP)2+ ion.
Histidine is a tridentate ligand having amino, imidazole and carboxylate groups as binding sites. With [Pd(DHP)(H2O)2]2+, only two of the three binding sites are involved in the complex formation, hence histidine coordinates in either a glycine-like or a histamine-like mode. The stability constant of the histidine complex is in fair agreement with that of histamine and higher than those of amino acids. This indicates that histidine interactes with the PdII complex in the same way as histamine coordinates. The proline complex has the highest value among the simple amino acids. This may be due to the highest basicity of the proline amino group as reflected by the highest pKa value. The stability constant with ornithine complex is higher than those of simple amino acids. This indicates that ornithine coordinates via the two nitrogen centres, i.e. by the two amino groups. This is in line with the strong affinity of PdII for nitrogen donor centres. The stability of methionine complex (log β = 8.78) is lower than those of most simple amino acids. This may be explained on the premise that the amino group of methionine is less basic than those of other amino acids as reflected by pKa values.
3.5 Complex formation equilibria involving peptides
The potentiometric data for the peptide complexes were fitted on the basis of formation of the complexes with stoichiometric coefficients 110 and 11-1, as given in Eq. (4)
| (4a) |
| (4b) |
The 110 complex is formed via coordination of the amine and carbonyl groups. On increasing pH, the amide groups undergo deprotonation associated with switch of coordination site from carbonyl to amide and the complex [Pd(DHP(LH−1)], (11-1) is formed. Such changes in coordination centres are well documented [26,27]. The pKH of the amide groups incorporated in the PdII complexes (log β110 − log β11-1) are in the 3.33–9.62 range. It is interesting to note that the pKa value for the glycinamide complex is lower than those for other peptides. This can be explained on the basis that the more bulky substituent on the peptide may hinder the structural changes when going from the protonated to the deprotonated complexes. The pKa of the glutamine complex is exceptionally higher than those for the other peptide complexes. This is due to the formation of a seven-membered chelate ring, which is more strained and therefore less favoured. The distribution diagram for the Pd(DHP)-glycylglycine system is given in Fig. 2. [Pd(DHP)L]+ (110), starts to form at lower pH, its concentration increases with increasing pH and reaches a maximum of 14% at pH 3.2. A further increase in pH is accompanied by a decrease in [Pd(DHP)L]+ (110) concentration and an increase in [Pd(DHP)LH-1] (11-1) concentration, reaching a maximum of 100% at pH 6.0, i.e. in the physiological pH range the deprotonated species (11-1) predominates.
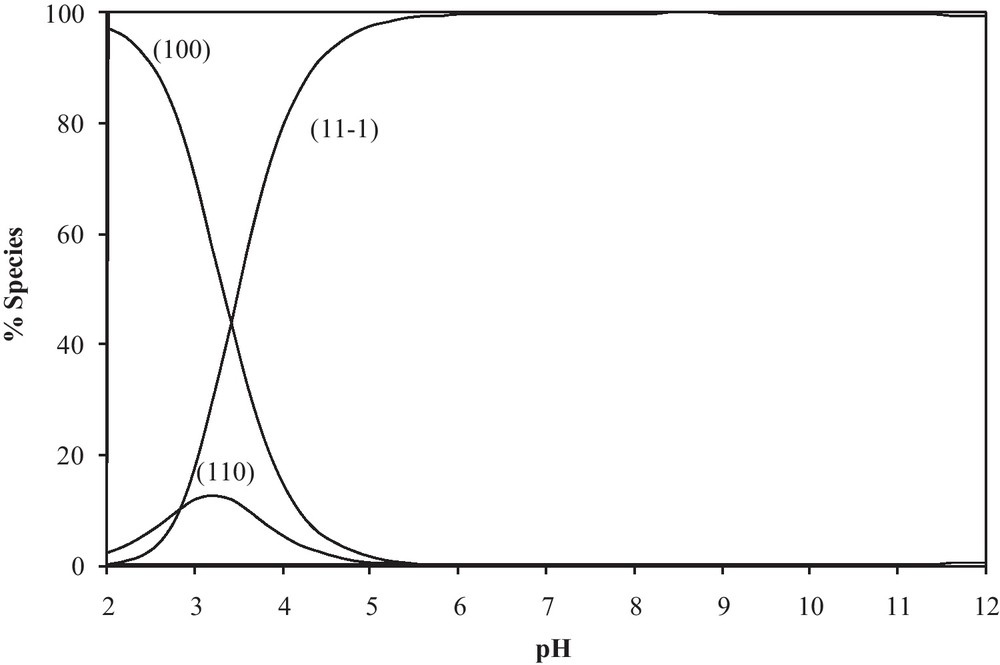
Concentration distribution curves for Pd(DHP)-glycylglycine complexes.
Spectral bands of Pd(DHP)(H2O)22+ and its glycylglycine complex are quite different in the position of the maximum wavelength and molar absorptivity (Fig. 3). The spectrum of Pd(DHP)(H2O)22+complex shows an absorption maximum at 354 nm. On the other hand the spectrum obtained for Pd(DHP)(glycylglycinate)+ (110 species), (mixture B), exhibits a band at 304 nm. The spectrum obtained for Pd(DHP)(glycylglycinateH−1) complex (11-1 species), (mixture C) exhibits a band at 282 nm. The progressive shift toward shorter wavelength in the absorption spectrum may be taken as evidence, supporting the potentiometric measurements for the induced ionization of amide upon complex formation.

The electronic spectra of: a: 1 mM of [Pd(DHP)(H2O)]2+; b: 1 mM of [Pd(DHP)(H2O)]2+, 1 mM of glycylglycine (HL) and 1 mM of NaOH; c: 1 mM of [Pd(DHP)(H2O)]2+, 1 mM of glycylglycine (HL) and 2 mM of NaOH.
3.6 Complex formation equilibria involving DNA constituents
The pyrimidines, uracil and thymine have only basic nitrogen donor atoms (N3-C4O group). They form 1:1 and 1:2 complexes with [Pd(DHP)(H2O)2]2+. Inosine is slightly more acidic than the pyrimidine bases, a property which can be related to the existence of a higher number of resonance forms for the inosine anion. Based on the existing data, uracil and thymine bind in the deprotonated form through the N3 atom. The thymine complex is more stable than that of uracil, most probably owing to the high basicity of the N3 group of thymine resulting from the extra-electron donating methyl group. As a result of the high pKa values of pyrimidines (pKa ≈ 9) and the fact that they are monodentate, the complexes are formed only above pH 6, supporting the view that the negatively charged nitrogen donors of pyrimidine bases are important binding sites in the neutral and slightly basic solution. The purines as guanosine 5’-monophosphate forms 110, 120, 111 and 112 complexes. The protonated species are formed in the acidic pH range and correspond to the N7 coordinated complex, where the N1 nitrogen and the phosphate group are protonated. The pKa values of the protonated species of the GMP complex (112) are 5.91 (log β112 − log β111) and 7.24 (log β111 − log β110). The former pKa value corresponds to the N1H group and the second pKa value to the −PO2(OH) group. The N1H group was acidified upon complex formation by 3.57 (9.48–5.91) pK units. Acidification of the N1H group upon complex formation is consistent with previous reports for IMP and GMP complexes [28]. The phosphate was not acidified upon complex formation since it is far away from the coordination centre.
3.7 Effect of solvent
It was reported that a lower polarity has been detected in some biochemical microenvironments, such as active sites of enzymes and side chains in proteins [29–33]. These properties approximately correspond to those (or can be simulated by those) existing in the water/dioxane mixtures. Consequently, a study of the Pd(DHP)-CBDCA complex formation, taken as a typical example, in dioxane-water solutions of different compositions could be of biological significance. In order to characterize the formation equilibria of the Pd(DHP)-CBDCA complex in dioxane-water solutions, all other equilibria involved, viz. acid-base equilibria of CBDCA and [Pd(DHP)(H2O)2]2+, have to be studied in the same solvent. The equilibrium constants are reported in Table 2. The hydrolysis of Pd(DHP)(H2O)2+ complex in dioxane-water solution leads to the formation of mono- and dihydroxy species. The dihydroxo-bridged dimer was not detected. The pKa values of CBDCA and those of the coordinated water molecules in [Pd(DHP)(H2O)2]2+ increase linearly with increasing of dioxane concentration. This may be correlated with the ability of a solvent of relatively low dielectric constant to increase the electrostatic attraction between the proton and ligand anion in case of CBDCA and that between a proton and the hydrolysed form of Pd(II) species. The variation in stability constant of the [Pd(DHP)(H2O)2]2+ complex with CBDCA as a function of solvent composition of range from 12.5 to 62.5% is investigated. The stability constant for the Pd(DHP)-CBDCA complex increases with increasing dioxane concentration. This is explained in terms of complex formation involving oppositely charged ions as in the Pd(DHP)-CBDCA complex, which is favoured by the low dielectric constant of the medium, i.e. with increasing dioxane concentration. The results show that the CBDCA complex with Pd(DHP)2+ will be more favoured in biological environments of lower dielectric constant.
Effect of dioxane on the formation constant of Pd(DHP)-CBDCA complex at 25 °C.
| % Dioxane | p | q | ra | log βb | Sc |
| 12.5 | 0 | 1 | 1 | 5.96 (0.02) | 5.7E-7 |
| 0 | 1 | 2 | 9.12 (0.04) | ||
| 1 | 1 | 0 | 7.58 (0.01) | 2.5E-7 | |
| 1 | 0 | −1 | −5.31 (0.02) | 1.2E-7 | |
| 1 | 0 | −2 | −15.18 (0.01) | ||
| 25 | 0 | 1 | 1 | 6.05 (0.01) | 1.5E-7 |
| 0 | 1 | 2 | 9.31 (0.02) | ||
| 1 | 1 | 0 | 7.84 (0.02) | 3.1E-7 | |
| 1 | 0 | −1 | −5.21 (0.01) | 2.5E-7 | |
| 1 | 0 | −2 | −15.04 (0.01) | ||
| 37.5 | 0 | 1 | 1 | 6.76 (0.01) | 1.1E-7 |
| 0 | 1 | 2 | 10.71 (0.01) | ||
| 1 | 1 | 0 | 8.79 (0.04) | 4.4E-7 | |
| 1 | 0 | −1 | −5.34 (0.05) | 3.8E-7 | |
| 1 | 0 | −2 | −15.25 (0.04) | ||
| 50 | 0 | 1 | 1 | 7.29 (0.01) | 1.7E-7 |
| 0 | 1 | 2 | 11.84 (0.02) | ||
| 1 | 1 | 0 | 9.78 (0.02) | 5.3E-7 | |
| 1 | 0 | −1 | −5.27 (0.02) | 5.0E-7 | |
| 1 | 0 | −2 | −15.26 (0.04) | ||
| 62.5 | 0 | 1 | 1 | 7.29 (0.01) | 1.4E-7 |
| 0 | 1 | 2 | 11.90 (0.02) | ||
| 1 | 1 | 0 | 9.79 (0.02) | 2.1E-7 | |
| 1 | 0 | −1 | −5.17 (0.05) | 2.5E-7 | |
| 1 | 0 | −2 | −15.27 (0.07) |
a p, q and r are the stoichiometric coefficients corresponding to Pd(DHP), CBDCA and H+ respectively.
b Standard deviations are given in parentheses.
c Sum of square of residuals.
3.8 Effect of chloride ion concentration on the equilibrium constants of [Pd(DHP)(H2O)]2+ complexes
Chloride ion competes with OH− for reaction with [Pd(DHP)(H2O)2]2+ complex, so the pKa values of the first and second coordinated water molecules increase with increasing chloride ion concentration and the results are included in Table 3. This means that the hydrolysis is suppressed by increasing of chloride ion concentration. The data of the effect of chloride ion concentration on the stability of [Pd(DHP)(CBDCA)] complex was given in Table 3. The stability constant of the 1:1 complex in Pd(DHP)-CBDCA system tends to decrease with increasing of [Cl−]. This is accounted for on the basis that the concentration of the active species, the diaqua- complex, decrease on increasing [Cl−], and this in turn will affect the stability of the complexes.
Effect Chloride Ion Concentration on the formation constant of Pd(DHP)-CBDCA complex at 25 °C and 0.30 M ionic strength.
| [Cl−] ion Concn. | p | q | ra | log βb | Sc |
| 0.00 M | 0 | 1 | 1 | 5.44 (0.01) | 1.9E-7 |
| 0 | 1 | 2 | 7.63 (0.04) | ||
| 1 | 0 | −1 | −5.20 (0.01) | 5.3E-8 | |
| 1 | 0 | −2 | −14.67 (0.01) | ||
| 1 | 1 | 0 | 7.10 (0.03) | 5.9E-6 | |
| 0.05 M | 1 | 0 | −1 | −7.41 (0.01) | 4.8E-8 |
| 1 | 0 | −2 | −16.99 (0.01) | ||
| 1 | 1 | 0 | 4.60 (0.02) | 4.3E-7 | |
| 0.10 M | 1 | 0 | −1 | −7.89 (0.01) | 8.2E-8 |
| 1 | 0 | −2 | −17.70 (0.01) | ||
| 1 | 1 | 0 | 4.11 (0.02) | 4.7E-7 | |
| 0.15 M | 1 | 0 | −1 | −8.28 (0.01) | 5.6E-8 |
| 1 | 0 | −2 | −18.10 (0.02) | ||
| 1 | 1 | 0 | 3.74 (0.04) | 2.9E-7 | |
| 0.20 M | 1 | 0 | −1 | −8.34 (0.01) | 8.3E-8 |
| 1 | 0 | −2 | −18.23 (0.01) | ||
| 1 | 1 | 0 | 3.53 (0.02) | 2.8E-7 | |
| 0.25 M | 1 | 0 | −1 | −8.51 (0.01) | 1.5E-7 |
| 1 | 0 | −2 | −18.45 (0.01) | ||
| 1 | 1 | 0 | 3.43 (0.03) | 6.0E-7 | |
| 0.30 M | 1 | 0 | −1 | −8.53 (0.01) | 1.5E-7 |
| 1 | 0 | −2 | −18.46 (0.03) | ||
| 1 | 1 | 0 | 3.37 (0.03) | 9.6E-7 |
a p, q and r are the stoichiometric coefficients corresponding to Pd(DHP), CBDCA and H+ respectively.
b Standard deviations are given in parentheses.
c Sum of square of residuals.
3.9 Displacement reaction of coordinated uracil
The preference of Pd(II) to coordinate to S-donor ligands was previously documented [26,27]. These results suggest that Pd(II)-N adducts can easily be converted into Pd-S adducts. Consequently, the equilibrium constant for such displacement reaction is of biological significance. Consider uracil as a typical DNA constituent (presented by HA) and cysteine as a typical thiol ligand (presented by H2B). The equilibria involved in complex-formation and displacement reactions are:
| (5a) |
| (5b) |
| (6a) |
| (6b) |
| (7a) |
| (7b) |
| (8) |
The equilibrium constant for the displacement reaction given in Eq. (8) is given by
| (9) |
Substitution from Eqs. (6b) and (7b) in Eq. (9) results in:
| (10) |
4 Conclusions
Pd(DHP)Cl2 and Pd(DHP)(CBDCA) complexes were synthesized and characterized. The interaction of [Pd(DHP)(H2O)2]2+ with ligands of biological significance was investigated. In combination of stability constants data of the complexes formed with dicarboxylic acids, amino acids, peptides and DNA constituents, it would be possible to calculate the equilibrium distribution of the Pd(II) species in biological fluids where all types of ligands are present simultaneously. This would form a clear basis for understanding the mode of action of such metal species under physiological conditions. The pKH of the amide groups incorporated in the PdII complexes has interesting biological implications. Under normal physiological conditions (pH 6-7) the peptide would coordinate to [Pd(DHP)(H2O)2]2+ in entirely different fashions. Glutaminate would exist solely in the protonated form, whereas the other peptides would be present entirely in the deprotonated form. In addition, the slight difference in the side chain of the peptides produces dramatic differences in their behaviour toward the palladium species. The equilibrium constant for displacement reaction of uracil by cysteine measures the deactivation of the Pt/Pd based-drug by the sulphur containing biomolecules.
A study of the Pd(DHP)-CBDCA complex stability, taken as a typical example, in dioxane-water solutions of different compositions could be of biological significance. The results show that the CBDCA complex with Pd(DHP)2+ will be more favoured in biological environments of lower dielectric constant.
Antitumour Pt(II)-amine complexes are usually administrated as cis-dichloro-complexes. This form persists in human blood plasma with its high 0.16 M Cl− ion content [34]. The net zero charge on the complex fasters its passage through cell walls. Within many cells the Cl− ion concentration is much lower, only 4 mM. Under this low chloride ion concentration, the reactivity of the Pt(II)-amine complex increases. Therefore a realistic extrapolation of the present study to biologically relevant conditions will require investigating the effect of [Cl−] on the stability constant of the complexes.


