1 Introduction
The field of molecular magnetism has attracted considerable attention, and advances have been made in designing novel molecular-based magnets with potential usage in information storage, and investigating the relationship between their structure and magnetic behavior [1]. Recently, special attention has been paid to the study of azido-bridging complexes in the field of molecular magnets [2–26]. The azide ligand is an extremely versatile ligand and also an excellent magnetic coupler, which gives easily tunable magnetic interactions between paramagnetic metal ions. This is due to the fact that the azide ligand can act as a bridge between transition metal atoms with different bridging coordination modes: μ-1,1 (end-on or EO) [2–7], μ-1,3 (end-to-end or EE) [7–15], μ-1,1,3 [16], μ-1,1,1 [3], and monodentate μ-1 [5,14], which typically obtain different magnetic coupling, such as antiferromagnetism [8,12–15], ferromagnetism [2,5], alternating ferro- and antiferromagnetism [17], ferrimagnetism [18], metamagnetism [19–21], spin-canting [22], spin-flop [23], spin-glass [24], single-molecule magnetism [25] and single-chain magnetism [26]. The magnetic coupling mediated through azide bridges is normally ferromagnetic [2,5] for the μ-1,1 (end-on) mode and antiferromagnetic for the μ-1,3 mode (end-to-end) [8,12–15]. Moreover, the magnetic properties for the μ-1,1 bridged metal complexes are dependent on the M–N–M angle, and for the μ-1,3 bridged metal complexes, it is dependent on the M–N–N angle and the M–N3–M dihedral angle [2,27].
To date, numerous azido-bridging metal complexes have been reported, including distinct polynuclear (dinuclear, trinuclear, and tetranuclear) [3,8,28], one-dimensional (1D) chain [2,12,15], two-dimensional (2D) layered [7,23] and three-dimensional (3D) framework complexes [5].
In continuation of our work on the azido-bridged complexes [29,30], herein, we report on the synthesis, structure and electrochemical properties of a new helical one-dimensional coordination polymer, [Mn(salabza)(μ-1,3-N3)]n, 1, and a centrosymmetric tetranuclear copper(II) coordination complex [Cu4(salabza)2(μ-1,1-N3)2(N3)2(HOCH3)2], 2 (Scheme 1). In complex 1, the azide ligand acts in end-to-end mode, and in complex 2, the azide ligand exhibits two types of coordination modes: end-on bridging fashion and terminal fashion.
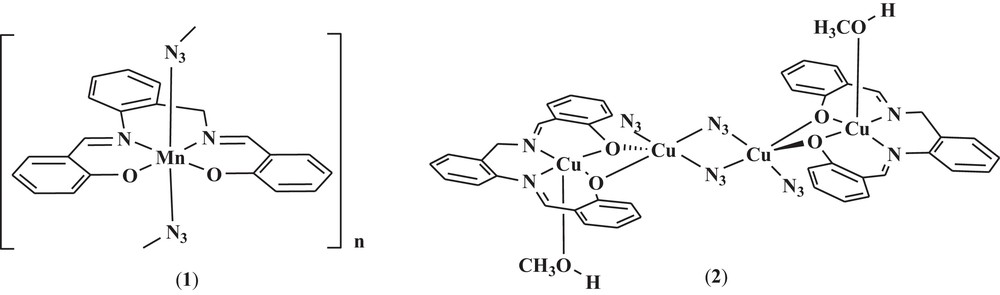
The molecular structures of the compounds reported in this paper.
2 Experimental
2.1 Materials and general methods
All the solvents and chemicals were of commercial reagent grade and used as received from Aldrich and Merck. Infrared spectra from KBr pellets were collected on a FT-IR JASCO 680 plus spectrometer in the range 4000–400 cm–1. UV–vis absorption spectra were recorded on a JASCO V-570 spectrophotometer. Elemental analyses were performed by using a PerkinElmer 2400 II CHNS–O elemental analyzer. Electrochemical properties of these complexes were studied by cyclic voltammetry. Cyclic voltammograms were recorded by using a SAMA 500 Research Analyzer using a three-electrode system, a glassy carbon working electrode (Metrohm 6.1204.110 with 2.0 ± 0.1 mm diameter), a platinum disk auxiliary electrode and an Ag wire as reference electrode. CV measurements were performed in DMF with tetrabutylammonium hexafluoro phosphate (TBAH) as the supporting electrolyte. The solutions were deoxygenated by purging with Ar for 5 min. All electrochemical potentials were calibrated versus an internal Fc+/0 (E0 = 0.46 V versus SCE) couple under the same conditions.
2.2 Synthesis of complexes
2.2.1 Synthesis of the [Mn(salabza)(μ-1,3-N3)]n complex (1)
To 10 mL of a methanolic solution of Mn(OAc)2·4H2O (0.049 g, 0.2 mmol) were added 0.066 g (0.2 mmol) of H2salabza [31] dissolved in 10 mL of chloroform, and the resulting solution was stirred for 30 min. A solution of NaN3 (0.052 g, 0.8 mmol) in methanol (l5 mL) was then added slowly to the above mixture. The resulting brown solution was allowed to stand in the open air for about 3 hours to complete the air oxidation of Mn(II). The final solution was left undisturbed at room temperature, and dark brown crystals appeared after 6 days. The crystals were collected by filtration, washed with small amounts of methanol and dried in the air (Yield: 64%) Anal. calc. for C21H16MnN5O2: C, 59.30; H, 3.79; N, 16.47. Found: C, 59.12; H, 3.68; N, 16.30%. FT-IR (KBr, cm–1) νmax: 1617, 1603 (CN), 2058 (bridging N3). UV–vis: λmax (nm) (ɛ, L mol–1 cm–1) (DMF): 322 (11942), 367 (9550), 620 (360).
2.2.2 Synthesis of the [Cu4(salabza)2(μ-1,1-N3)2(N3)2(HOCH3)2] complex (2)
The complex 2 was prepared by a procedure similar to that of 1 except that Cu(OAc)2.H2O (0.04 g, 0.2 mmol) was used instead of Mn(OAc)2·4H2O. Dark brown crystals were collected by filtration and washed with small amounts of methanol (Yield: 72%). Anal. calc. for C44H40Cu4N16O6: C, 46.31; H, 3.36; N, 19.64. Found: C, 47.31; H, 3.18; N, 19.17%. FT-IR (KBr, cm–1) νmax: 1630, 1608 (CN), 2035 (bridging N3), 2072 (terminal N3). UV–vis: λmax (nm) (ɛ, L mol–1 cm–1) (DMF): 384 (32 036), 622 (860).
2.3 Crystal structure determination and refinement
2.3.1 X-ray crystallography for 1 and 2
Single crystals of 1 and 2 were obtained by slow evaporation of methanol–chloroform (1:1 v/v) solutions of these complexes at room temperature.
X-ray data of these compounds were collected at T = 100 K on a Bruker Kappa APEXII CCD diffractometer with graphite monochromated Mo Kα (λ = 0.71073 Å) radiation. Cell refinement and data reduction were performed with program SAINT [32]. Correction for absorption was carried out by the multi-scan method and program SADABS [32]. The structure was solved using the program SHELXS97 and structure refinement on F2 was carried out with the program SHELXL97 [33]. Structural diagrams were created with program DIAMOND [34].
3 Results and discussion
3.1 Description of structures
3.1.1 Crystal structure of [Mn(salabza)(μ-1,3-N3)]n (1)
The diagram of a single unit with its atomic labeling scheme and the one-dimensional polymeric structure of [Mn(salabza)(μ-1,3-N3)]n are shown in Figs. 1 and 2, respectively. The crystallographic and refinement data for 1 are summarized in Table 1, and selected bond distances and angles are listed in Table 2.
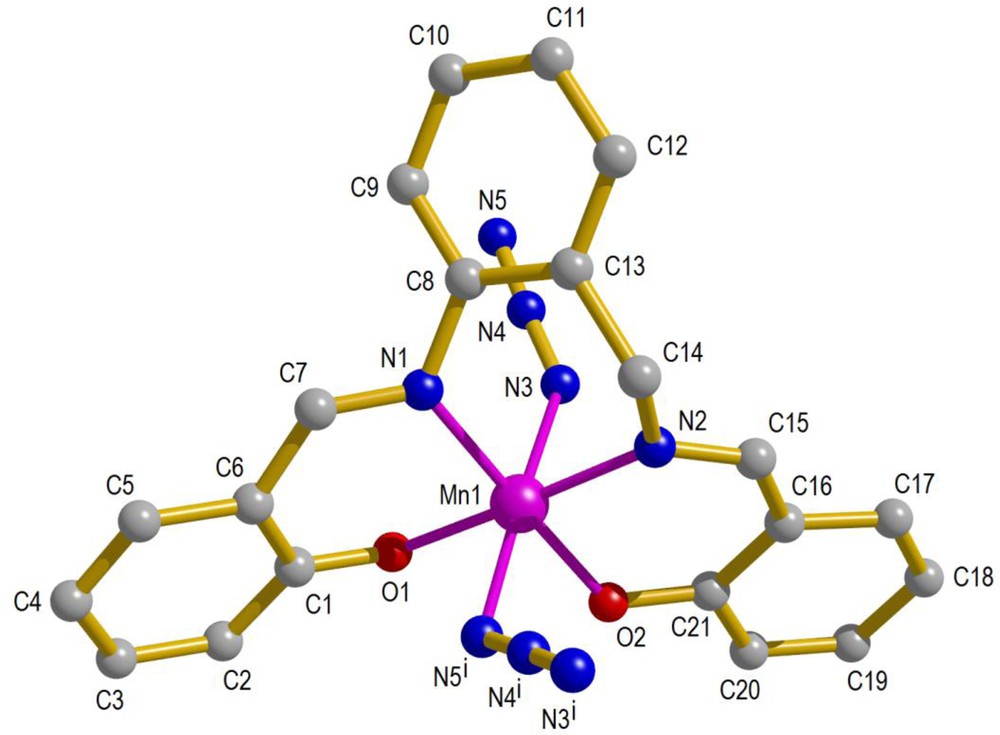
The molecular structure of 1 with its atom labeling scheme. Symmetry code: (i) 1 – x, ½ + y, ½ – z. For interpretation of colors, see the online version of this article.
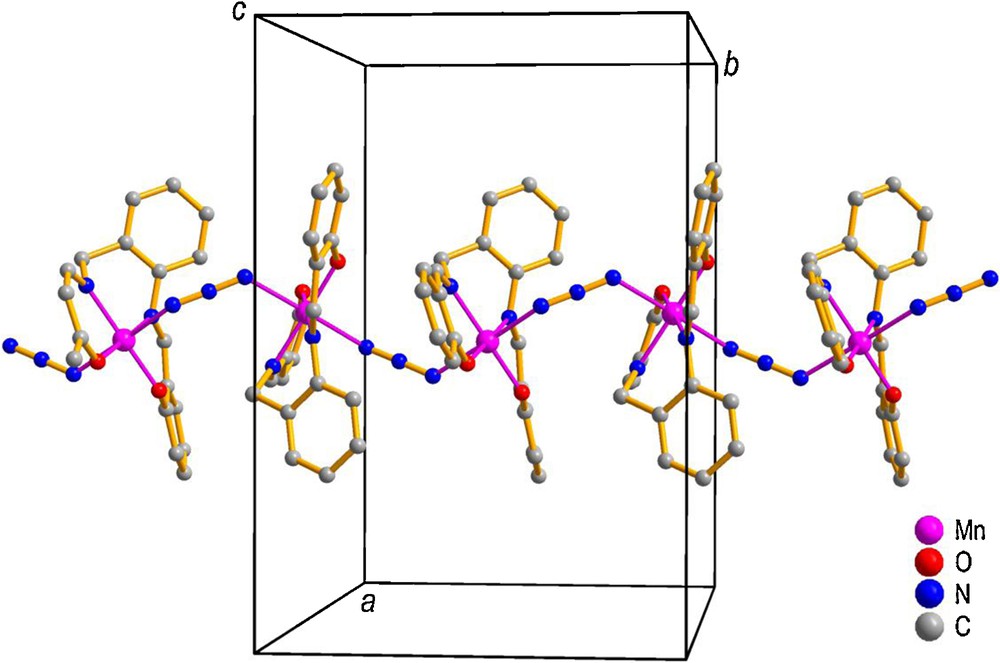
The structure of 1, showing the helical 1D-polymeric chain extending along the b axis. For interpretation of colors, see the online version of this article.
Crystal data and structure refinement for 1 and 2 complexes.
| Compound | (1) | (2) |
| Empirical formula | C21H16MnN5O2 | C44H40Cu4N16O6·2(CH4O) |
| Formula weight | 425.33 | 1207.16 |
| Temperature (K) | 100 (2) | 100 (2) |
| Crystal system | Orthorhombic | Triclinic |
| Space group | Pbca | Pī |
| a (Å) | 16.5827 (5) | 9.8597 (4) |
| b (Å) | 11.2289 (3) | 11.0946 (5) |
| c (Å) | 19.8004 (5) | 13.3326 (9) |
| α (°) | 90.00 | 99.216 (3) |
| β (°) | 90.00 | 107.350 (3) |
| γ (°) | 90.00 | 112.957 (2) |
| V (Å3) | 3686.94 (18) | 1216.76 (11) |
| Z | 8 | 1 |
| Dcalc (mg/m3) | 1.532 | 1.647 |
| μ (mm–1) | 0.75 | 1.80 |
| Crystal size (mm) | 0.52 × 0.18 × 0.16 | 0.48 × 0.32 × 0.28 |
| F (000) | 1744 | 616 |
| θ Ranges (°) | 2.1–30.1 | 2.1–30.0 |
| Index ranges | −2 ≤ h ≤ 23 | −13 ≤ h ≤ 13 |
| −15 ≤ k ≤ 15 | −15 ≤ k ≤ 15 | |
| −27 ≤ l ≤ 27 | −18 ≤ l ≤ 18 | |
| Absorption correction | Multi-scan | Multi-scan |
| Reflections collected | 77879 | 30846 |
| Rint | 0.034 | 0.025 |
| Data/restraints/parameters | 5415/0/262 | 7058/0/337 |
| Goodness-of-fit on F2 | 1.02 | 1.06 |
| Final R indices [I > 2σ(I)]a | R1 = 0.027, wR2 = 0.072 | R1 = 0.035, wR2 = 0.088 |
| Largest diff. peak and hole (e·Å–3) | −0.23 and 0.49 | −0.80 and 1.56 |
a R1 = ∑||Fo| – |Fc||/ ∑|Fo|, wR2 = {∑[w(Fo2 – Fc2)2]/ ∑[w(Fo2)2]}1/2.
Selected bond lengths (Å) and angles (°) for (1).
| Mn1—O1 | 1.9003 (8) | Mn1—N3 | 2.2155 (10) |
| Mn1—O2 | 1.8922 (8) | Mn1—N5i | 2.2453 (11) |
| Mn1—N1 | 2.0243 (10) | N3—N4 | 1.1730 (15) |
| Mn1—N2 | 2.0278 (9) | N4—N5 | 1.1734 (15) |
| O1—Mn1—O2 | 92.91 (3) | O2—Mn1— N5i | 90.60 (4) |
| O1—Mn1—N1 | 88.07 (4) | N1—Mn1—N2 | 88.55 (4) |
| O1—Mn1—N2 | 176.07 (4) | N1—Mn1—N3 | 91.06 (4) |
| O1—Mn1—N3 | 94.67 (4) | N1—Mn1—N5i | 90.56 (4) |
| O1—Mn1—N5i | 86.86 (4) | N2—Mn1—N3 | 87.40 (4) |
| O2—Mn1—N1 | 178.52 (4) | N2—Mn1—N5i | 91.16 (4) |
| O2—Mn1—N2 | 90.52 (4) | N3—Mn1—N5i | 177.81 (4) |
| O2—Mn1—N3 | 87.76 (4) | Mn1—N3—N4 | 140.98 (9) |
| Mn1—N5i—N4i | 127.40 (9) |
[Mn(salabza)(μ-1,3-N3)]n crystallizes in the orthorhombic space group Pbca. In this complex, each Mn atom is in octahedral environment, consisting of N2O2 donor atoms (two phenoxo oxygen and two imine nitrogen atoms) from the salabza ligand in equatorial positions and two nitrogen atoms from two bridging azide groups in axial positions. The Mn(salabza) units are linked by the azide bridging groups, in μ-end-to-end (EE) mode, to form a one-dimensional coordination polymer. The unique feature of the resulting chain is that each monomeric unit is related to its neighboring ones by a 2-fold screw axis, leading to a helix propagating along the crystallographic b axis. The equatorial donor atoms N1, N2, O1, and O2 are nearly co-planar, with slight deviation from the mean plane: 0.026, –0.026, –0.027, and 0.027 Å, respectively, and the Mn atom deviates by only 0.007 Å from this plane. As expected for the Mn(III) ion, the axial Mn–N3 and Mn–N5 distances (2.2155 (10) and 2.2453 (11) Å, respectively) are longer than the two Mn–N distances in equatorial sites (Mn–N1 = 2.0243 (10) Å and Mn–N2 = 2.0278 (9) Å), due to the Jahn–Teller distortion of Mn(III) ions. In this complex, the bond angles about Mn are slightly distorted from the ideal values for a regular octahedral structure. The N–Mn–N cis angles are in the range 87.40 (4)–91.16 (4)° and the N–Mn–O trans angles are 176.07 (4) and 178.52 (4)°. All distances and angles of this complex are consistent with the corresponding distances in related manganese(III) complexes [15,22,35].
3.1.2 Crystal structure of [Cu4(salabza)2(μ-1,1-N3)2(N3)2(HOCH3)2] (2)
The molecular structure of complex 2 is shown in Fig. 3. The crystallographic and refinement data for 2 are summarized in Table 1, and selected bond distances and angles are listed in Table 3.
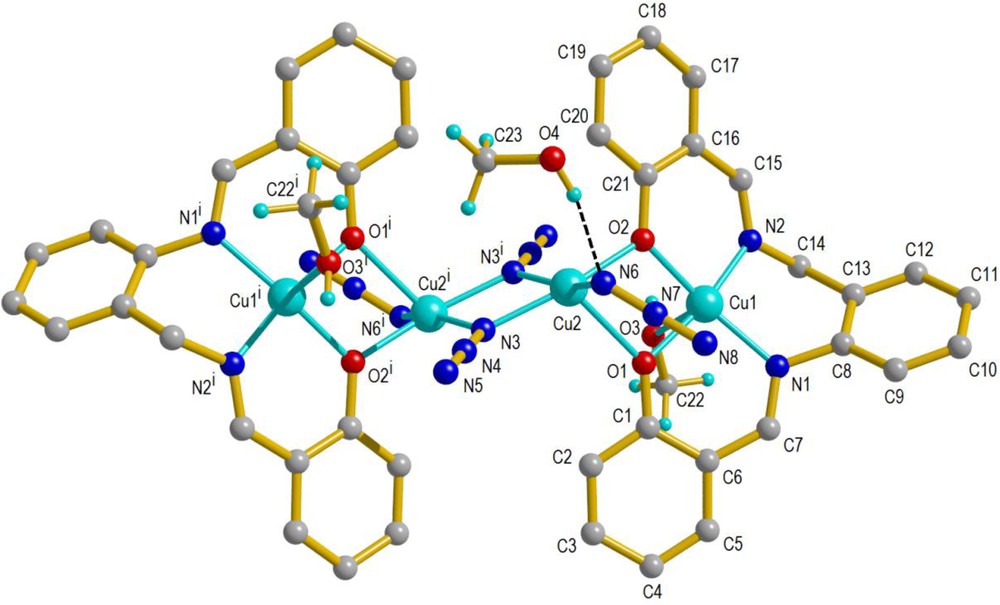
The molecular structure of 2 with its atom labeling scheme. Symmetry code: (i) 1 – x, 1 – y, 1 – z. For interpretation of colors, see the online version of this article.
Selected bond lengths (Å) and angles (°) for (2).
| Cu1—O1 | 1.9100 (13) | Cu2—O1 | 2.2728 (13) |
| Cu1—O2 | 1.9393 (13) | Cu2—O2 | 1.9795 (13) |
| Cu1—O3 | 2.4569 (17) | Cu2—N3 | 1.9865 (16) |
| Cu1—N1 | 1.9692 (16) | Cu2—N3i | 2.0062 (16) |
| Cu1—N2 | 1.9399 (16) | Cu2—N6 | 1.9637 (17) |
| O1—Cu1—O2 | 81.86 (6) | O1—Cu2—O2 | 72.38 (5) |
| O1—Cu1—O3 | 84.69 (6) | O1—Cu2—N3 | 111.54 (6) |
| O1—Cu1—N1 | 94.41 (6) | O1—Cu2—N3i | 104.79 (6) |
| O1—Cu1—N2 | 168.55 (7) | O1—Cu2—N6 | 94.37 (6) |
| O2—Cu1—O3 | 100.81 (6) | O2—Cu2—N3 | 168.29 (6) |
| O2—Cu1—N1 | 159.23 (7) | O2—Cu2—N3i | 91.17 (6) |
| O2—Cu1—N2 | 92.31 (6) | O2—Cu2—N6 | 96.74 (7) |
| O3—Cu1—N1 | 99.16 (7) | N3—Cu2—N3i | 77.20 (7) |
| O3—Cu1—N2 | 86.76 (6) | N3—Cu2—N6 | 93.97 (7) |
| N1—Cu1—N2 | 94.46 (7) | N3i—Cu2—N6 | 160.72 (7) |
| Cu1—O1—Cu2 | 97.99 (5) | N4—N3—Cu2 | 128.50 (14) |
| Cu1—O2—Cu2 | 107.75 (6) | N4—N3—Cu2i | 127.66 (14) |
| Cu2—N3—Cu2i | 102.81 (7) | N7—N6—Cu2 | 118.04 (15) |
This complex crystallizes in triclinic space group , and the structure consists of a centrosymmetric tetranuclear molecule with two types of Cu(II) centers. The coordination geometry around all copper atoms is distorted square pyramidal. The terminal copper(II) ions, Cu1, are coordinated with N2O2 donor atoms from the salabza ligand in basal positions and one oxygen atom from CH3OH in apical position. The central copper(II) ions, Cu2, are coordinated with one of the two phenoxo oxygen atoms from one salabza ligand, one terminal azido ligand, and two end-on (EO) bridging azido groups in the basal plane. The axial position is occupied by the second phenoxo oxygen atom from the same salabza ligand. The central copper(II) ions are linked to each other via two end-on (EO) azido groups.
In Cu1 centers, the bond lengths in the basal plane are Cu1–N1 = 1.9692 (16), Cu1–N2 = 1.9399 (16), Cu1–O1 = 1.9100(13), Cu1–O2 = 1.9393 (13) Å, and the axial Cu1–O3methanol distance (2.4569 (17) Å) is markedly longer than the basal Cu1–Ophenoxo distance. This axial elongation could be attributed to the Jahn–Teller distortion of copper(II) ions. The deviations of the four basal donor atoms from the mean basal plane N1, N2, O1, and O2 are –0.223, 0.227, 0.256, and –0.261 Å, respectively, and the Cu1 atom is 0.098 Å out of the plane towards the apex. The two trans angles around the Cu1 centers, O1–Cu1–N2 = 168.55 (7)° and O2–Cu1–N1 = 159.23 (7)°, deviate significantly from the ideal value of 180° for a regular square pyramidal structure. The trigonality index (Addison parameter) τ = 0.155 {τ = (β – α)/60°, where α, β are the two largest L–M–L angles of the coordination sphere, with τ = 0 and 1 for perfect square pyramidal and trigonal bipyramid geometries, respectively [36]} confirms the distorted square pyramidal environment for Cu1.
In Cu2 centers, the basal plane around the Cu is defined by N3, N3i, N6 and O2, with bond lengths of Cu2–N3 = 1.9865 (16), Cu2–N3i = 2.0062 (16), Cu2–N6 = 1.9637 (17), and Cu2–O2 = 1.9795 (13) Å. The apical position of Cu2 is occupied by O1 with the Cu2–O1 bond length 2.2728 (13) Å, which is different from the Cu2–O2 bond length in basal position. This axial elongation is due to the Jahn–Teller effect reflecting the unique character of the copper(II) ion. The deviations of the four basal donor atoms from the mean basal plane N3, N3i, N6, and O2 are 0.132, –0.134, –0.111, and 0.113 Å, respectively, and the Cu2 atom is 0.176 Å out of the plane towards the apex. The two trans angles, O2–Cu2–N3 = 168.29 (6)° and N6–Cu2–N3i = 160.72 (7)°, deviate significantly from the ideal value. The trigonality index of Cu2 is τ = 0.126, while the corresponding value for the terminal copper(II) atoms is 0.155, indicating that the geometry of the terminal copper(II) atoms is more distorted than that of the central one. All distances and angles of this complex are consistent with the corresponding distances in related copper(II) azide complexes [28,37–39].
The intramolecular Cu···Cu distances between the adjacent copper atoms in 2 are 3.166 Å for Cu1···Cu2 and 3.121 Å for·Cu2···Cu2i. Intermolecular hydrogen bond interactions occur between the hydroxyl H atoms of the methanol molecules (coordinated as well as uncoordinated) and the terminal N atoms of the azide groups (Table 4). The hydrogen bonds of the Cu1-bonded methanol, O3—H3O···N5, link the molecules in infinite chains parallel to [100] (Fig. 4). The uncoordinated methanol molecules establish branches.
Hydrogen bonds for 2 (Å and°).
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3O···N5ii | 0.84 | 2.14 | 2.930 (3) | 156 |
| O4—H4O···N6 | 0.84 | 2.16 | 2.972 (3) | 162 |
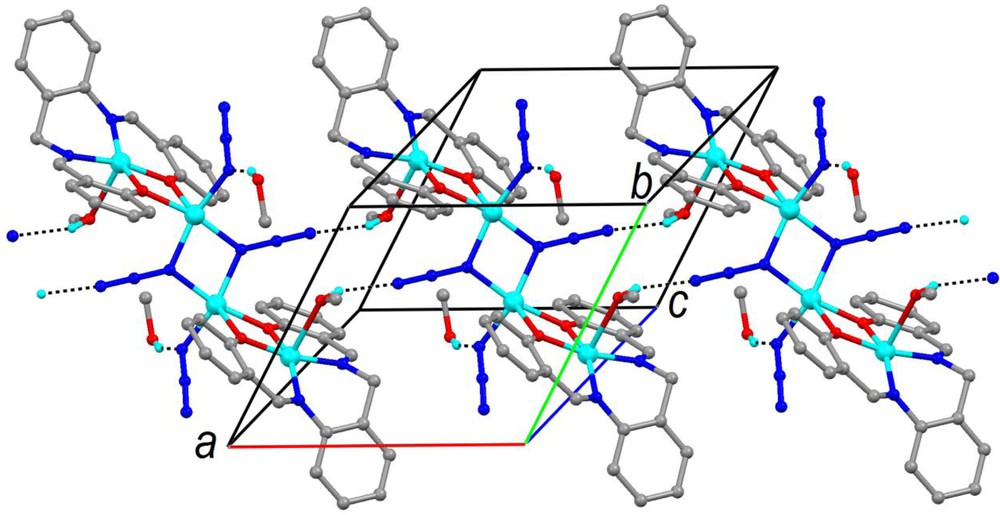
Packing and hydrogen bonds (dashed lines) of 2 C-bonded H atoms were omitted for clarity. For interpretation of colors, see the online version of this article.
3.2 Spectral characterization
The FT-IR spectral data of the two complexes are listed in Section 2. The vibrations appearing at 1617 and 1603 cm–1 for 1 and at 1630 and 1608 cm–1 for 2 are characteristic of two different imine (CN) groups, as expected for the unsymmetrical Schiff base ligands. The strong absorption band at 2058 cm–1 in 1 is assigned to the bridging azido ligand, and the two strong absorption bands at 2035 and 2072 cm–1 in 2 clearly indicate the presence of both bridging and terminal azide ligands.
The UV–visible spectra of these compounds were recorded in DMF solution in the 200–800 nm region and the data are presented in Section 2. The absorption spectrum of Mn(III) complex consisted of a weak band at 620 nm attributed to the metal-centered 5Eg → 5T2g transition, and two relatively intense bands at 367 and 322 nm corresponding to intraligand and charge-transfer transitions. The copper(II) complex shows a weak band at 622 nm, assigned to the d–d transition, 2B1 → 2A1, and an intense band at 384 nm, corresponding to the charge-transfer transition. The intraligand transition is observed as a shoulder close to the charge-transfer band.
3.3 Electrochemical studies
The cyclic voltammogram of [Mn(salabza)(μ-1,3-N3)]n in DMF solution (Fig. 5) shows a quasi-reversible reduction process at –0.185 V, which is attributed to the reduction of MnIII/MnII. A possible explanation of this feature is that in DMF solution compound 1 transforms into a mononuclear complex [Mn(salabza)(κ1-N3)(κ1O-DMF)] [40].
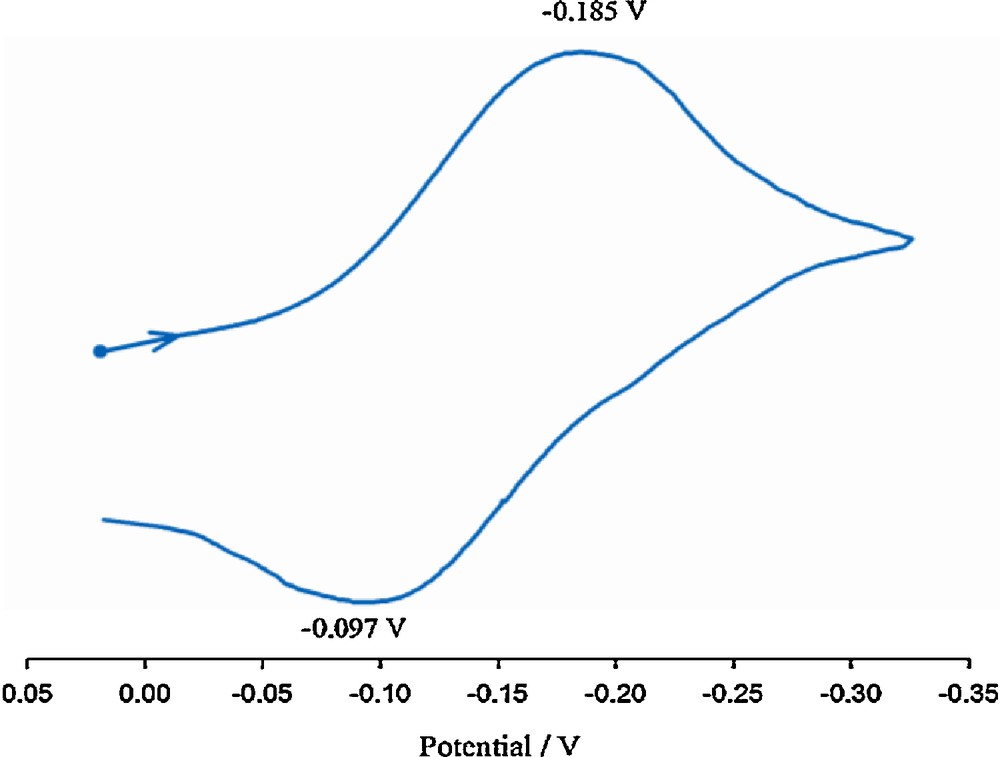
Cyclic voltammogram of the 1 in DMF at 298 K, c ≈ 1 × 10–3 M, scan rate = 100 mV/s.
The cyclic voltammogram of the [Cu4(salabza)2(μ-1,1-N3)2(N3)2(HOCH3)2] complex in DMF is shown in Fig. 6. Two successive quasi-reversible two-electron reductions were observed at 0.105 V for two Cu2 centers and at –1.055 V for two Cu1 centers. These CuII/CuI reductions reflect the existence of two types of Cu(II) centers with different coordination environments. The redox process for Cu2 centers appear at more positive potential as compared to that for the corresponding Cu1 centers, due to the electron-withdrawing nature of the azido ligands.
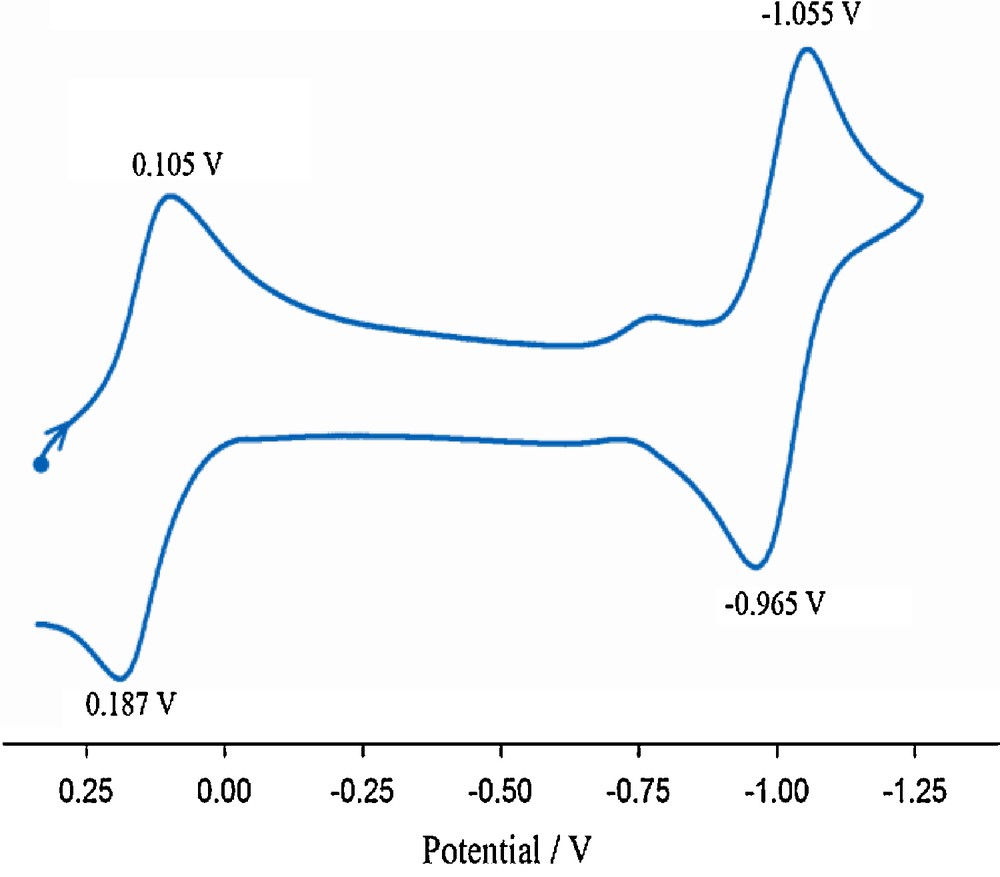
Cyclic voltammogram of the 2 in DMF at 298 K, c ≈ 1 × 10–3 M, scan rate = 100 mV/s.
4 Conclusions
In summary, a new one-dimensional azido-bridged manganese(III) polymer, [Mn(salabza)(μ-1,3-N3)]n, and a centrosymmetric tetranuclear azido-bridged copper(II) complex [Cu4(salabza)2(μ-1,1-N3)2(N3)2(HOCH3)2] have been synthesized, and fully characterized The single crystal X-ray structure analysis of these complexes shows that, in complex 1, the azide ligand acts in end-to-end (μ-1,3) mode, and in complex 2, the azide ligand exhibit two types of coordination modes: end-on bridging (μ-1,1) mode and terminal mode.
Acknowledgements
Partial support of this work by the Isfahan University of Technology Research Council is gratefully acknowledged.


