1. Introduction
The double-helical structure of DNA in which two antiparallel strands are held together through canonical A/T and G/C base pairing was established over half a century ago. However, the last decades brought accumulating evidence of the existence and biological relevance of four-stranded nucleic acid structures namely G-quadruplex (G4) and i-motif (i-DNA). G4 structures could be formed from guanine rich sequences and consist in stacked tetrads of Hoogsteen hydrogen-bonded guanine nucleobases (i.e. G-tetrad or quartet, Figure 1A), connected by various loop-forming sequences, and stabilized through the coordination of physiologically abundant cations (Na+, K+) [1, 2].
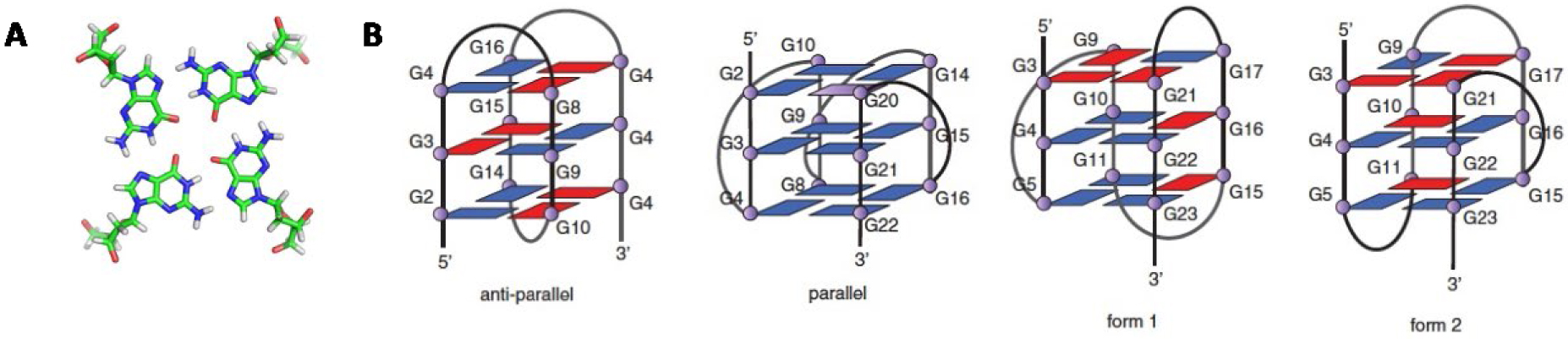
(A) G-tetrad (or G-quartet), (B) schematic representations of some G-quadruplex topologies with three G-tetrads from reference [2].
Bioinformatics studies suggested that the human genome contains around 370,000 sequences having the potential to form stable G-quadruplex structures (PQS) [3]. This was reevaluated using a novel algorithm and high-resolution sequencing-based method (termed “G4-seq”), which identified more than 700,000 PQS within the genome [4, 5]. Interestingly, these putative G4s are not distributed randomly in the genome. Indeed a statistically significant enrichment of PQS was found in several relevant domains of the genome. DNA G4-forming sequences can be found in the telomeric region where their stabilization has been shown to inhibit activity of telomerase, which is over-expressed in 80% of cancer cells, thus evidencing their potential as anticancer drug targets [6, 7]. However G4 formation is not limited to the telomeric region: they are also over-represented in the promoter regions of a number of genes, including proto-oncogenes c-Myc, c-Kit, bcl-2 and KRAS [8]. Furthermore, the majority of the 250,000 human replication origins are close to G4 motifs suggesting that the formation of stable G4 structures participates in the initiation of replication [9]. It has been reported that certain pathologies or chronic diseases caused by cell dysfunction might involve the presence of G4. G4 formation has been linked to genetic disorders (diabetes, fragile X disorder, Bloom syndrome), age-related degenerative illness (ALS, FTD) and cancer (telomere, MYC, Kit, BCL-2). G4 formation has also been evidenced in the genomes of viruses suggesting functional significance [10]. Besides G4-DNA, G-rich RNA sequences are also prone to fold into stable G4 architectures (G4-RNA) [11]. G4-RNA-forming sequences can be found in the 5′- and 3′-untranslated regions of many genes, and also in the open reading frame of some mRNAs [11]. To date, the formation of G4-RNA has been involved in several biological processes linked to RNA metabolism such as translation regulation, pre-mRNA processing, and mRNA targeting. Owing to the single-stranded nature of transcribed RNA, in vivo formation of G4-RNA is expected to occur more easily than G4-DNA. Strong arguments have been provided that argue in favor of the formation of DNA and RNA G4 structures within cells, by using G4-specific antibodies [12], in vivo NMR [13], and binding-activated fluorescent G4-targeting ligands [14].
An essential feature of G4 is their intrinsic polymorphic nature: numerous in vitro studies have revealed their susceptibility to adopt different topologies, which are in equilibrium. Indeed, depending on the length and composition of the sequence, as well as the environmental conditions (including the nature and concentration of metal cations, and local molecular crowding), a G-quadruplex-forming sequence can adopt different topologies in which the strands are in parallel or antiparallel conformations, with the co-existence of different types of loops (lateral, diagonal or propeller) with variable lengths (Figure 1B) [1].
Cytosine, the complementary nucleobase of guanine, is also prone to assemble to form four-stranded structures named i-motif DNA (i-DNA) in which cytosines are intercalated via a stack of hemi-protonated (CH+:C) base pairs (Figure 2) [15].
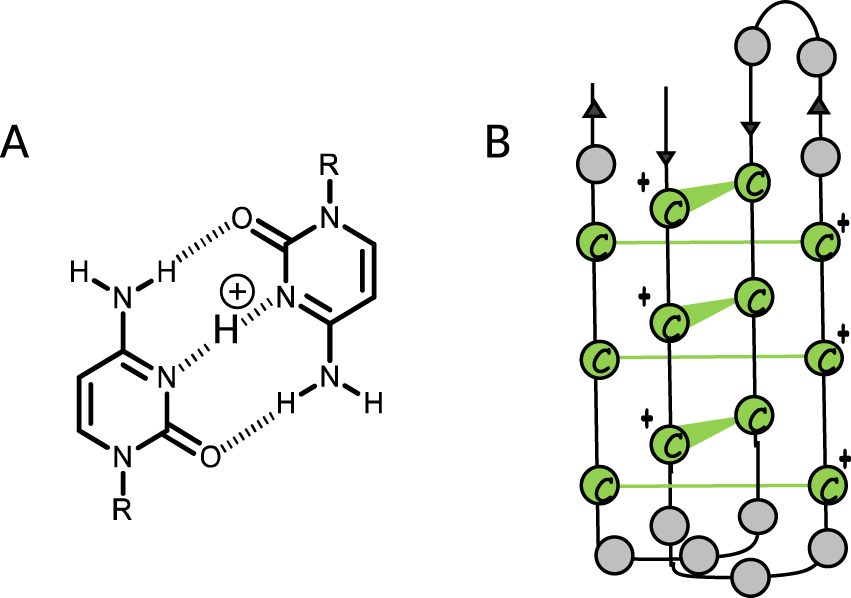
(A) (CH+:C) base pairing, (B) schematic representation of i-DNA.
A major characteristic of i-DNA is the strong pH-dependency of its stability and formation. Indeed, i-DNA structures are typically observed in vitro at acidic pH, a particularity that has cast doubt on their existence in cell. However, two independent studies have recently demonstrated the stability of exogenous i-DNA structures in human cells through in cellulo NMR spectroscopy [16] as well as the presence of endogenous i-DNA in the nuclei of human cells through immunofluorescence using an antibody (i-Mab) raised against the i-motif [17].
The biological relevance of i-DNA has been less investigated mainly due to the scientific community’s skepticism about the existence of i-motifs in vivo. A recent review from Brown and Kendrick provides some insight into the biological function of i-DNA structure [18]. In the context of bcl-2 oncogene, a transcriptional activation was reported, caused by the formation of stable i-motif through the interaction with IMC-48 compound, a cholestane derivative (selected from a screening of a NCI diversity set library) [19]. In contrast with bcl-2 oncogene, a transcriptional repression was reported with c-MYC i-motif [20]. Also, proteins such as hnRNP LL have been identified to interact with i-motif structures acting as an activating transcription factor [21].
The intrinsic polymorphism associated with the formation of G4 and i-DNA as well as the pH-dependency of i-DNA stability represent severe bottlenecks for the studies of those tetrameric DNA structures. Indeed, the polymorphism could lead to intricate structural mixtures in solution that can complicate the rationalization of the relationships between G4 or i-motif structures and recognition by proteins and ligands. Likewise, low-pH conditions used to induce the formation of i-DNA could lead to the protonation of many ligands (e.g., proteins), strongly increasing their non-specific nucleic acid binding. The design of chemical tools able to reduce the structural heterogeneity of G4 and i-DNA as well as able to improve i-motif stability in physiological conditions is thus of high interest.
In this context, we developed some years ago an innovative concept that consists to constrain the accessible topologies of a G-quadruplex-forming sequence to a single one [22]. This strategy named TASQ for Template-Assembled Synthetic Quadruplex, is based on the use of a rigid cyclic peptide scaffold with two independently functionalizable faces, resulting from the orientation of the lysine side-chains. One face is dedicated to the anchoring of different oligonucleotide sequences to obtain the desired G4 topology and a biotin residue is incorporated on the other side for attachment to streptavidin immobilized surfaces for various applications (Figure 3). This template concept allowed the formation of very stable G-quadruplex motifs in a unique conformation, in aqueous medium. It was next extended to the formation of constrained i-motif DNA.
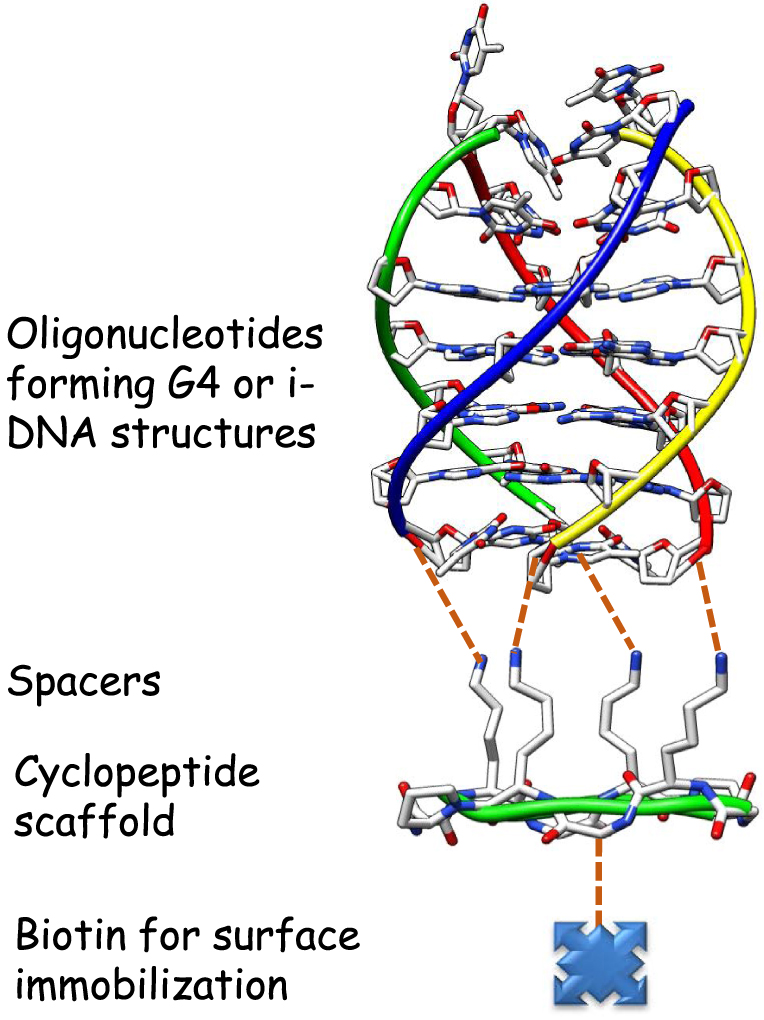
Schematic representation of the TASQ concept. The different oligonucleotide sequences forming the target structure are attached to the cyclopeptide scaffold through different ligation techniques (see below) on the top of the cyclopeptide and a biotin residue is incorporated in the lower face of the cyclopeptide.
In this article, we describe our contribution for the design of efficient chemical tools based on constrained nucleic acids for the study of G4 and i-motif DNA. Different applications and perspectives of those chemical tools are then described.
2. Results and discussions
2.1. Synthesis of the various constrained G-quadruplex and i-DNA systems
The design of constrained DNA in the chemical biology research domain has already been investigated. As an example, Escudier et al. have developed modified oligonucleotides in which the phosphodiester internucleotidic linkage is replaced with a dioxo-1,3,2-oxaza-phosphorinane moiety resulting in conformationally constrained nucleotides (CNA) [23]. In the case of G-quadruplex, the use of various templates to pre-organize G-quartet assemblies has also been described by different groups [24, 25, 26, 27].
Our approach consisted in the use of cyclic peptide scaffolds based on the TASP concept (Template-Assembled Synthetic Proteins) for the design of folded proteins developed by Mutter in 1985 [28]. The chemoselectively addressable cyclic peptide template is the key intermediate as it exhibits two independent and chemically addressable domains, which allows the sequential and regioselective assembly of the different oligonucleotides forming the tetrameric nucleic acid target on one face, the other face serving for the attachment on surfaces. By using sequential ligation techniques we were able to prepare different G-quadruplex systems (Figure 4).
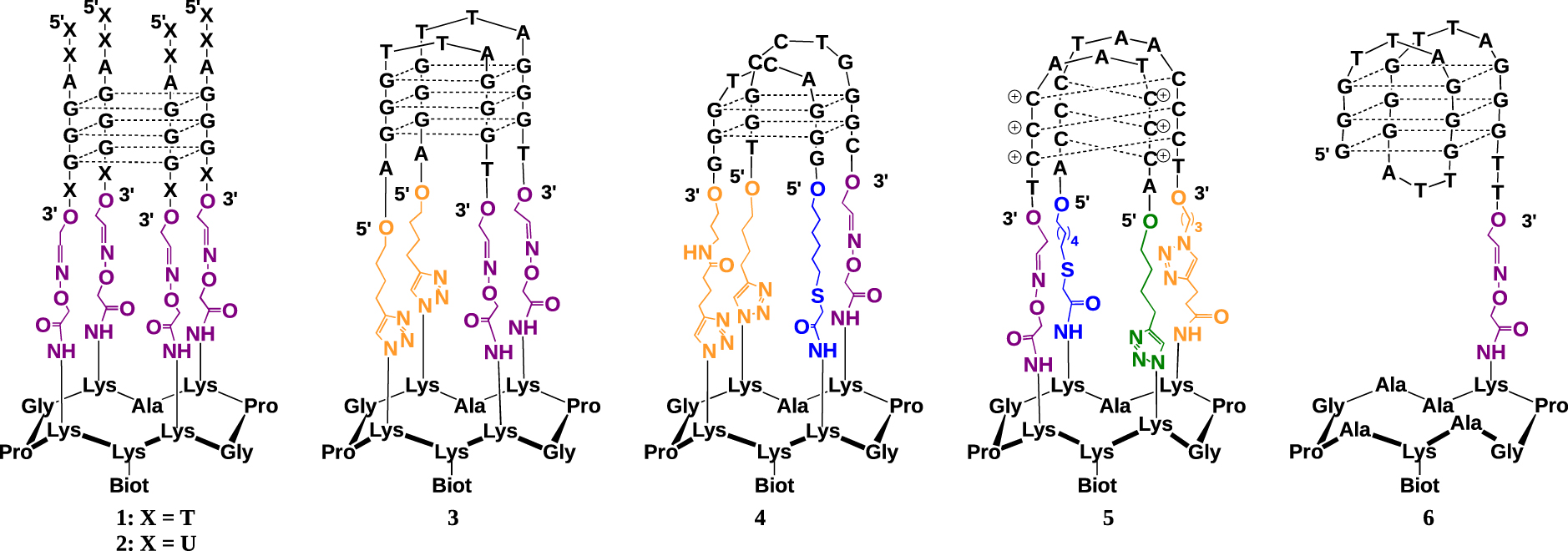
Structure of the different constrained tetrameric nucleic acid structures 1–5 and unconstrained control 6.
Conjugate 1 was the first constrained G4 prepared [22]. It may mimic intermolecular-like G-quadruplexes. Four oligonucleotides derived from human telomeric sequence d(5′-TTAGGGT-3′) were attached onto the peptide scaffold by using oxime bond formation from 3′-aldehyde-containing oligonucleotides and the peptide scaffold bearing four aminooxy residues. By using CD melting studies, we demonstrated that the peptide template allows the formation of a very stable G4 motif in a unique parallel conformation, in aqueous medium. Using the same oxime ligation (OL) method, we later synthetized the corresponding RNA G42 from telomeric sequence (TERRA) and found again that the template allows stabilization of the desired parallel topology [29].
The synthesis of antiparallel topology required the use of two successive ligation reactions for the attachment of oligonucleotides at both 3′ and 5′ extremities. This was achieved through sequential oxime (OL) and Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAc) reactions. The antiparallel topologies of G-quadruplex DNA from telomeric sequence 3 was obtained by the reaction of 3′-aldehyde, 5′-alkyne bis-functionalized oligonucleotides with the suitable cyclopeptide and was shown to exhibit high stability and reduced polymorphism [30].
Next, we expanded the TASQ approach to stabilize biologically relevant viral G4 structures such as the one found in sequence (5′-TGGCCTGGGCGGGACTGGG-3′) derived from the LTR region of HIV-1 [31]. Unlike with telomeric G4-forming conjugates 1-3, the site-specific attachment onto the cyclopeptide scaffold of the two G-rich DNA oligonucleotides with sequences 5′-TGGCCTGGGC-3′ and 5′-GGACTGGG-3′, respectively, mimicking the sequence from LTR region of HIV-1, implied the use of an additional orthogonal chemical ligation step along with OL and CuAAC. This was achieved using a SN2-thiol coupling reaction (TC). Conjugate 4 was thus prepared through successive conjugations, first with a functionalized oligonucleotide bearing an aldehyde at its 3′-end and an alkyne at its 5′-end, then with another one bearing a thiol function at its 5′-end and an alkyne at its 3′-end [32].
Lastly, the construction of conjugate 5, a mimic of i-motif DNA formed from the telomeric sequence, was carried out. The synthesis of the i-motif structural mimic 5 was achieved via the stepwise assembly of peptide–DNA conjugates through four successive ligations with one OL, one TC and two CuAAC reactions [33]. The resulting conjugate 5 was found by CD to fold, at room temperature, into an i-motif structure which is stable at acidic and neutral pH. It may therefore be used to study, at physiologically relevant pH, the interaction of the i-motif with putative i-motif targeting ligands (i.e., small molecules or proteins).
2.2. Applications of the constrained DNA chemical tools
The different constrained G-quadruplex and i-motif DNA 1-5 were used to study previously described ligands or new molecules able to interact with the G4 or i-DNA targets. The main objectives of those studies were to investigate the interactions of the ligands with different DNA targets and to assess their selectivity versus other DNA structures as well as versus different G-quadruplex topologies.
With these chemical tools in our hands, another goal was to identify, by classical capture methods, and characterize proteins which bind to a predetermined single G4 topology, and study their interactions with the diverse structural motifs (i.e., loops, grooves, quartets) of the quadruplexes by comparing their binding properties to different defined constructs. This application is now extended to i-DNA.
We also envision to use the constrained G-quadruplex to produce and characterize antibodies for a given G4 topology by using the above-described chemical tools. We believe that preventing the equilibrium between the different conformations that are associated with G-quadruplex-forming sequences will facilitate the production of specific antibodies. Again this will be extended to i-DNA.
2.3. Use of constrained systems to study the interactions with ligands
Most of the knowledge of the impact of G4-DNA secondary structures on cell metabolism resulted from the use of selective chemical probes that bind or modulate the formation of such structures [34]. A major challenge in G-quadruplex ligand synthesis is the development of compounds that are able to distinguish G-quadruplexes from duplex DNA and also discriminate between various G4 topologies. A detailed picture of quadruplex structure is emerging from crystallographic and NMR studies, and together with computer modeling, it is possible to develop a rational approach toward the design and optimization of quadruplex-stabilizing compounds [35]. The desirable features of these stabilizing molecules are (i) a π-delocalized system that is able to stack on the face of a guanine quartet; (ii) a partial positive charge that lies in the center of the quartet, increasing stabilization by substituting for the cationic charge of the potassium or sodium that would normally occupy that site; and (iii) positively charged substituents that will interact with the grooves and loops of the quadruplex and the negatively charged backbone phosphates.
In contrast to G4 ligands, relatively few molecules were reported to interact with i-DNA, and a controversy concerning their binding mode, affinity, and selectivity persists in the literature [36, 37]. The main challenges in this regard are the strong pH-dependency, flexibility and polymorphism of i-DNA, introducing potential bias into screening methods. Indeed, low-pH conditions used to induce the formation of i-DNA lead to the protonation of many ligands, strongly increasing their non-specific nucleic acid binding.
To investigate the interactions of our constrained tetrameric nucleic acids with potential ligands, two optical techniques were used: surface plasmon resonance (SPR) and bio-layer interferometry (BLI). Those two label-free techniques are widely used to study the interactions of ligands (including proteins, nucleic acids, sugars, and small molecules) with analytes. The ligand is immobilized on the surface while the analyte is injected close to the surface via a micro-fluidic system for SPR or deposited in microplate for BLI. The sensorgram fittings provide the association and dissociation kinetic constants, and the responses obtained at the steady state (Req) afford the equilibrium dissociation constant (KD). Those two techniques display a number of advantages, including the non-use of special radioactive or fluorescent labeling of the molecules, the time efficiency, the use of very low quantity of materials associated with a high sensitivity, the access to a variety of commercial surface sensors and the possibility to assemble homemade sensors bearing specific chemical functionalities. Possible drawbacks of BLI/SPR techniques are related to the relatively high cost of such equipment as well as the requirement of a good expertise (i.e., to not over/mis-interpret the results).
2.3.1. Study of the interaction with well-known G-quadruplex ligands
A large number of G-quadruplex ligands have been reported in the literature and most of them interact with G-quadruplex DNA by π-stacking interactions with the external G-quartet of the quadruplex [38, 39]. We have used some of them with the aim of verifying if the constrained G-quadruplex systems could act as efficient mimics before the investigation of unknown ligands. Moreover, we envisioned that our different G-quadruplex systems could afford some information about the mode of interaction. The following reported ligands TMPyP4 [40], MMQ1 [41], distamycin [42], FRHR [43], and DODC [44] were first studied (Figure 5). As anticipated, ligands displaying a π-stacking binding mode such as TMPyP4 showed a higher binding affinity for intermolecular-like G-quadruplex 1 due to the absence of loops which could prevent the interactions, whereas ligands with other binding modes (groove and/or loop binding) such as distamycin showed no significant difference in their binding affinities for the constrained quadruplex 1 and unconstrained control 6 [45]. In addition, the method has also provided information about the selectivity of ligands for G-quadruplex DNA over the duplex DNA through comparative studies with DNA hairpin duplex. Further studies with other well-known G-quadruplex ligands such as Phen-DC3 [46], PDS [47], BRACO-19 [48] and NMM [49] (Figure 5) were carried out next. The use of constrained or not constrained G4 systems also allowed to obtain some information about the selectivity for the ligands with a single G4 topology. Most of the described ligands do not show any G4 topology preference, except NMM. We have demonstrated the high selectivity of NMM for the parallel G4 structure with a dissociation constant at least ten times lower than those of other G4 topologies as well as the ability of this ligand to shift the G4 conformation from both the hybrid and antiparallel topologies toward the parallel structure [50].
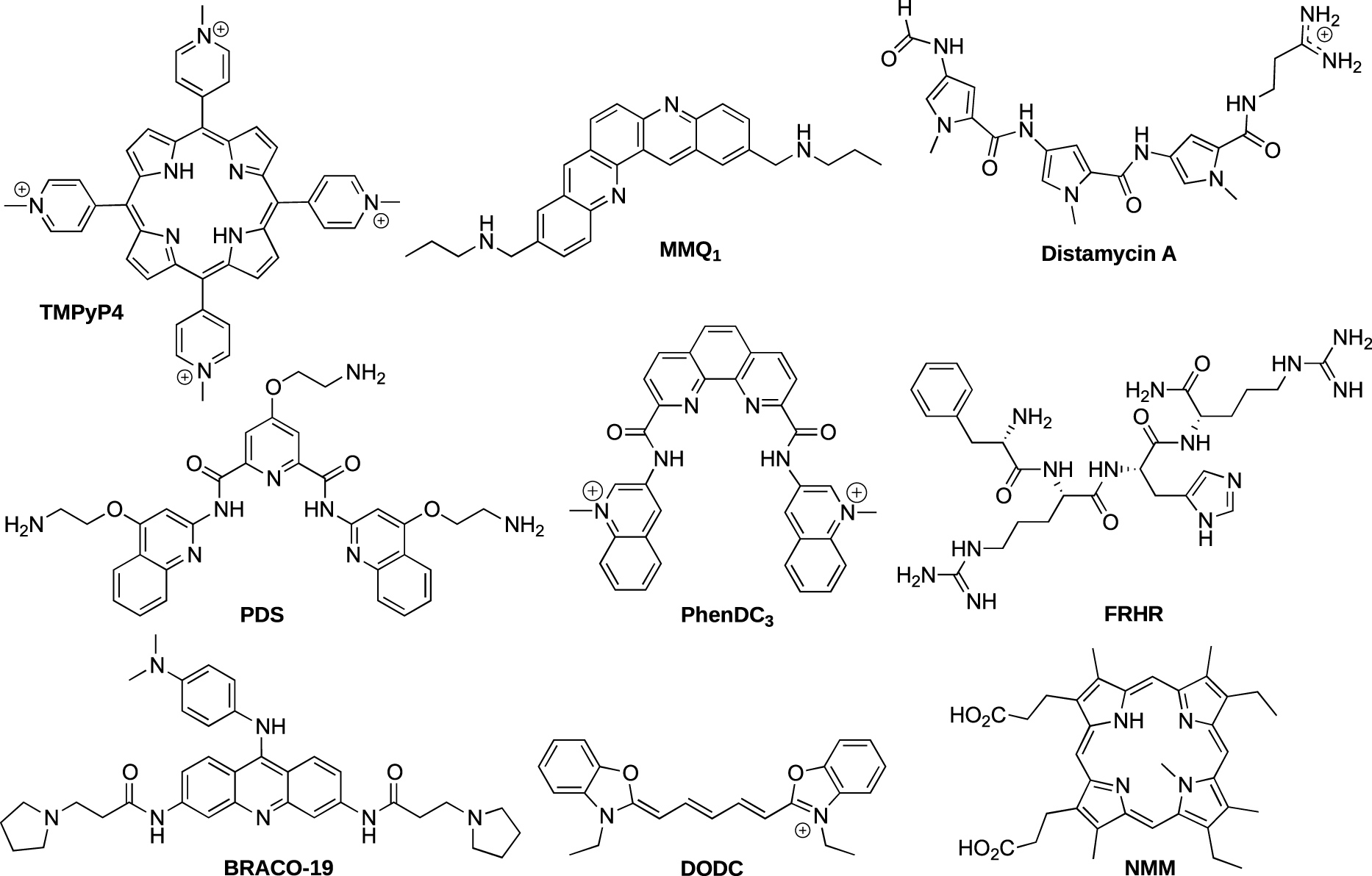
Structure of the different reported ligands.
The studies with well-known G-quadruplex ligands thus validated that the constrained systems are useful tools for investigating the interactions with G-quadruplexes. With those chemical tools in hands, we next investigated the interactions with various ligands designed by our collaborators.
2.3.2. Study of the interaction of different families of G-quadruplex ligands based on metal complexes
In the field of targeting G-quadruplex nucleic acids structures with small molecules, hundreds of ligands have been reported [38, 39]. Most of them interact with G-quadruplex DNA by π-stacking interactions with the external G-quartet of the quadruplex. The design of metal complexes targeting G-quadruplex DNA has also attracted intense interest [51]. In comparison to organic compounds, metal complexes show many advantages, such as a net positive charge (i.e., able to increase the interactions with DNA), tunable geometry, and, most interestingly, some of them display potentially useful photochemical properties. In that context, we were interested in the design and study of different classes of G-quadruplex binders based on metal complexes including metal porphyrin derivatives, salophens, and ruthenium and iridium photoreactive complexes.
2.3.2.1. Porphyrin-based ligands.
One of the first reported G-quadruplex ligands was the non-metalated porphyrin, meso-5,10,15,20-tetrakis(4-N-methylpyridiniumyl)-porphyrin (TMPyP4, Figure 5). The main disadvantage of TMPyP4 is its weak selectivity for G4 versus all other DNA structure. In the aim of improving selectivity, Pratviel et al. have designed new porphyrin derivatives through the insertion of a metal ion (Ni2+, Co3+, or Mn3+) into the porphyrin core (TMPyP4 series 8–10) or by modification of the meso substituents R of the porphyrin with a phenyl-N-methylpyridinium group (TMPyP4-PP series 11–14) and a guanidinium group (TMPyP4-PG 15–17) as depicted in Figure 6 [52, 53, 54].
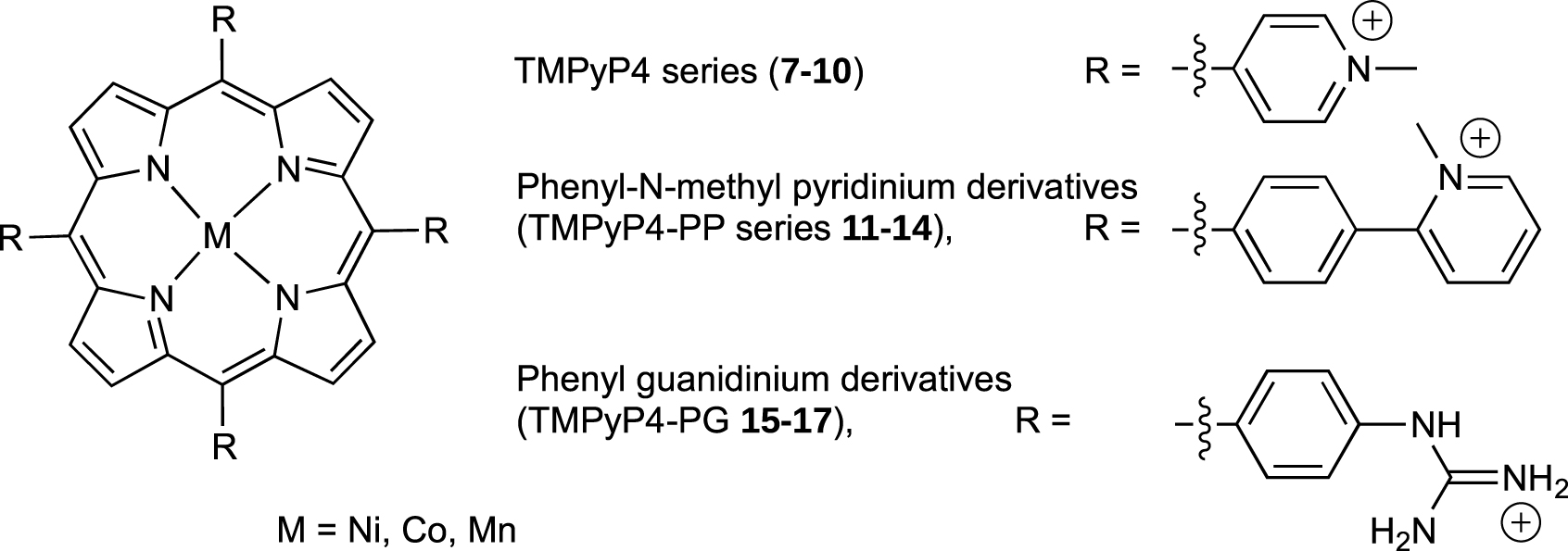
Structure of metalated TMPyP4 derivatives. The un-metalated ligands TMPyP4 7, TMPyPP4-PP 11 and TMPyP4-PG 15 correspond to those structures without M.
By using the constrained G4 system 1, we have demonstrated that the insertion of a metal ion into TMPyP4 (Ni, Co or Mn) or modification of the porphyrin meso substituents could drastically modify affinity and selectivity for the G4. For example TMPyP4 derivatives 9 and 10 with cobalt and manganese metal respectively, showed a high selectivity for G-quadruplexes 1 and 6 versus duplex DNA as no interaction occurs with duplex DNA, whereas for TMPyP4 derivative 8 with nickel metal as well as for parent TMPyP4 7 an interaction for duplex DNA quite equivalent to that for G-quadruplex 6 was observed (Figure 7). That was because water/hydroxo as axial ligands on the cobalt and manganese derivatives could preclude the intercalation of the porphyrin moiety between the base pairs of duplex. For TMPyP4-PP 11–14 and TMPyP4-PG 15–17 series, the presence of bulky substituents should also prevent the intercalation between the base pairs of duplex DNA leading to a weak interaction of those compounds with duplex DNA in comparison with quadruplexes 1 and 6 (Figure 7).
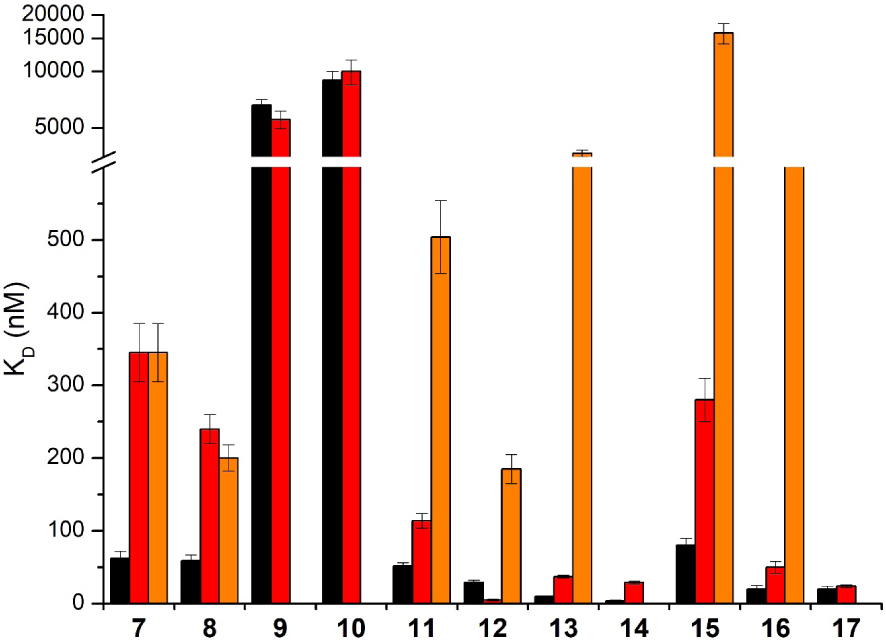
KD values obtained with the different porphyrin derivatives 7–17 in interaction with G-quadruplexes 1 (black), unconstrained control 6 (red) and duplex DNA (orange). 7: TMPyP4, 8: Ni-TMPyP4, 9: Co-TMPyP4, 10: Mn-TMPyP4, 11: TMPyP4-PP, 12: Ni-TMPyP4-PP, 13: Co-TMPyP4-PP, 14: Mn-TMPyP4-PP, 15: TMPyP4-PG, 16: Ni-TMPyP4-PG, 17: Mn-TMPyP4-PG. No histogram for duplex DNA means that the KD value could not be determined due to too weak interaction. The reported values are the means of representative independent experiments, and the errors provided are standard deviations from the mean. Each experiment was repeated at least three times. Masquer
KD values obtained with the different porphyrin derivatives 7–17 in interaction with G-quadruplexes 1 (black), unconstrained control 6 (red) and duplex DNA (orange). 7: TMPyP4, 8: Ni-TMPyP4, 9: Co-TMPyP4, 10: Mn-TMPyP4, 11: TMPyP4-PP, 12: Ni-TMPyP4-PP, 13: Co-TMPyP4-PP, 14: ... Lire la suite
Guanidinium phenyl porphyrin derivatives 15–17 exhibit moderate cytotoxicity toward cells in culture. Strikingly, the nickel porphyrin derivative 15 was able to displace hPOT1 sheltering protein from telomeres in human cells [53].
2.3.2.2. Ni-Salphen derivatives
Pioneering works by Neidle et al. revealed that salphen derivatives bind strongly to the human telomeric G-quadruplexes and inhibit the telomerase activity with EC50 of roughly 0.1 μM [55]. In order to study the impact on binding affinity of the length of the side-chains and their positions on the salphen scaffold, Thomas et al. have prepared new family of G-quadruplex binders based on the nickel(II) salphen platform (Figure 8). The side-chains are alkyl-imidazolium arms connected at para, ortho or meta positions of the phenol moieties [56, 57]. The affinity for G-quadruplex DNA 1 and 6 as well as the selectivity versus duplex DNA were evaluated by using SPR.
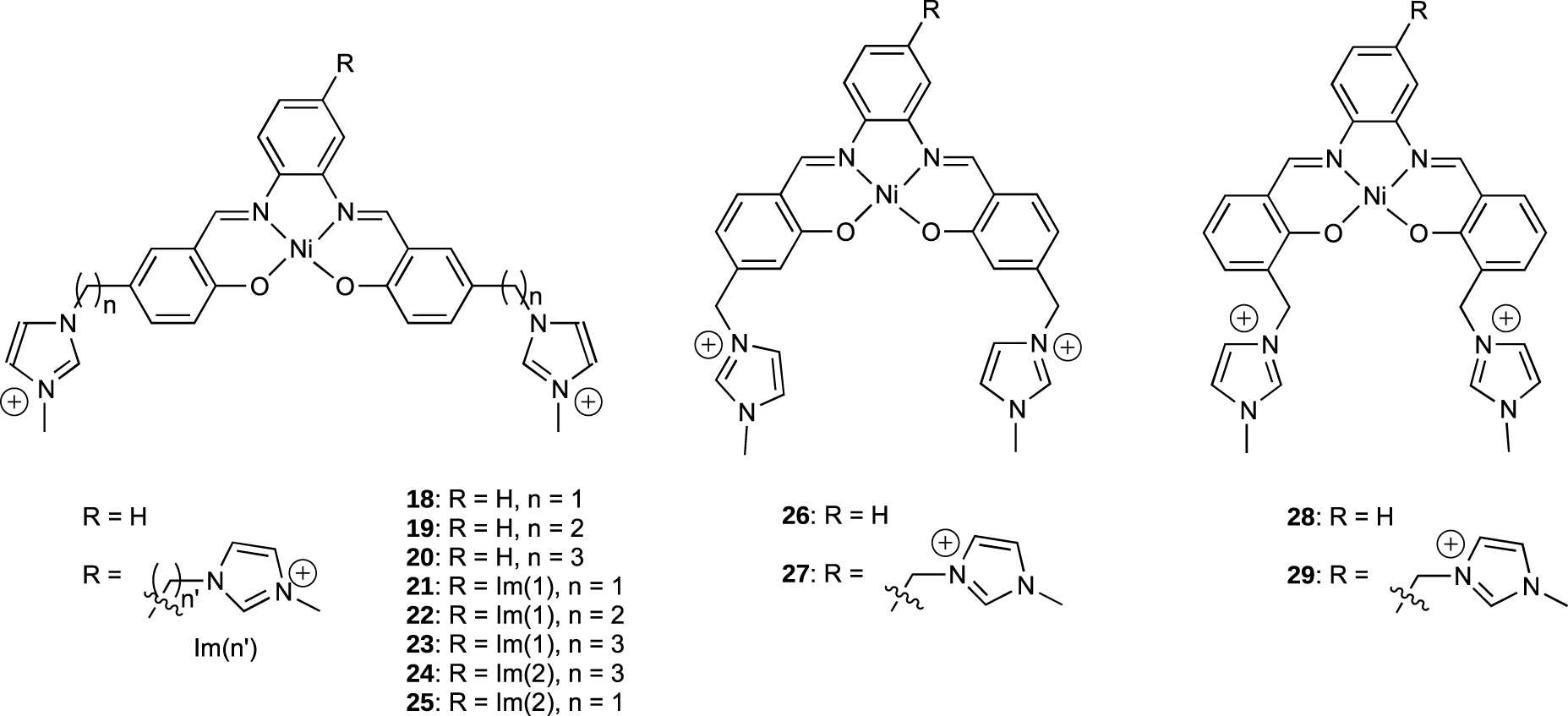
Structure of the different Ni-Salphen derivatives.
The different salphen derivatives 18–29 showed KD values in the 0.1–2 μM range for both G-quadruplex 1 and 6, which are in the range of those reported for related compounds interacting with the HTelo sequence [58] and most of them do not bind tightly to duplex DNA (Figure 9).
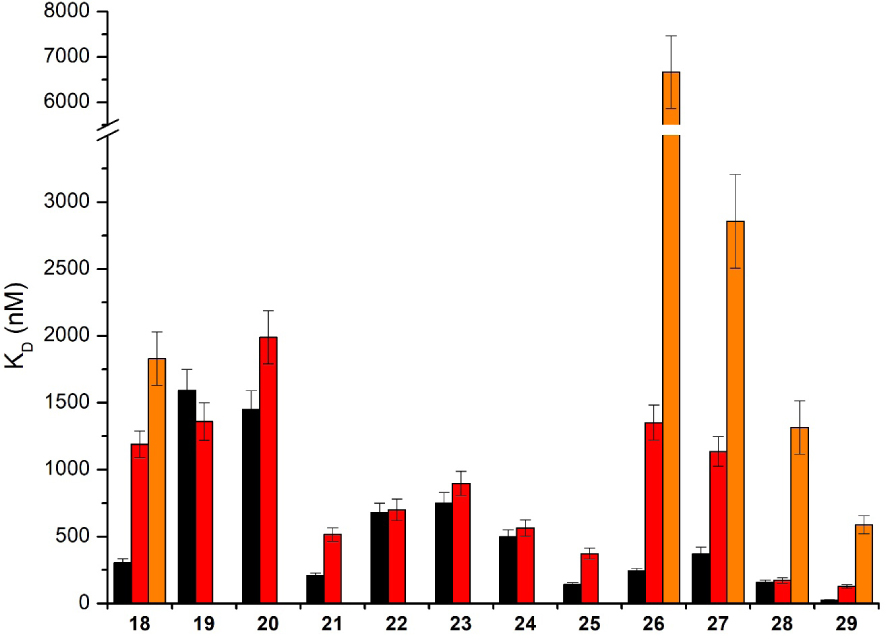
KD values obtained with the Ni-salphen derivatives 18–29 in interaction with G-quadruplexes 1 (black), 6 (red) and duplex DNA (orange). No histogram for duplex DNA means that the KD value could not be determined due to too weak interaction. The reported values are the means of representative independent experiments, and the errors provided are standard deviations from the mean. Each experiment was repeated at least three times.
SPR studies with our systems 1 and 6 allowed to obtain some useful information. We observed that the shorter the side arms, the higher the affinity for G4. Furthermore the introduction of a third anchor on the diaminobenzene bridge also improved the affinity for G4. The difference of affinity for 1 versus 6 also suggested that the compounds interact both by π-stacking over the tetrad and electrostatic interactions in the grooves. From the SPR studies, we have concluded that salphen 21, 25 and 29 were the optimal G-quadruplex binders: they were also the best salphen derivatives able to inhibit telomerase activity and in particular with an IC50 value measured from TRAP-G4 assays of 70 nM for 29.
2.3.2.3. Ruthenium and iridium complexes
Ruthenium(II) and iridium metal complexes have been investigated as G4-ligands and some of them have shown good affinity and selectivity toward G4s [51, 59, 60, 61]. In this context, Elias et al. have developed several ruthenium and iridium complexes which are able to target G-quadruplex DNA (Figure 10). The affinity for G-quadruplexes and the selectivity versus DNA duplexes were investigated using BLI and SPR (Figure 11).
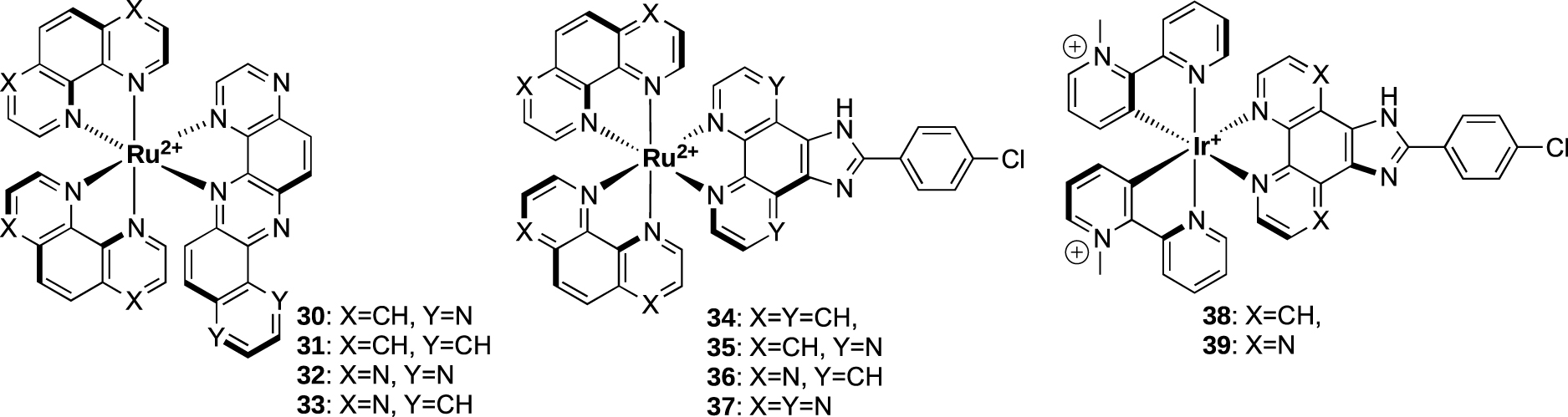
Structure of ruthenium and iridium complexes from Elias collaboration.
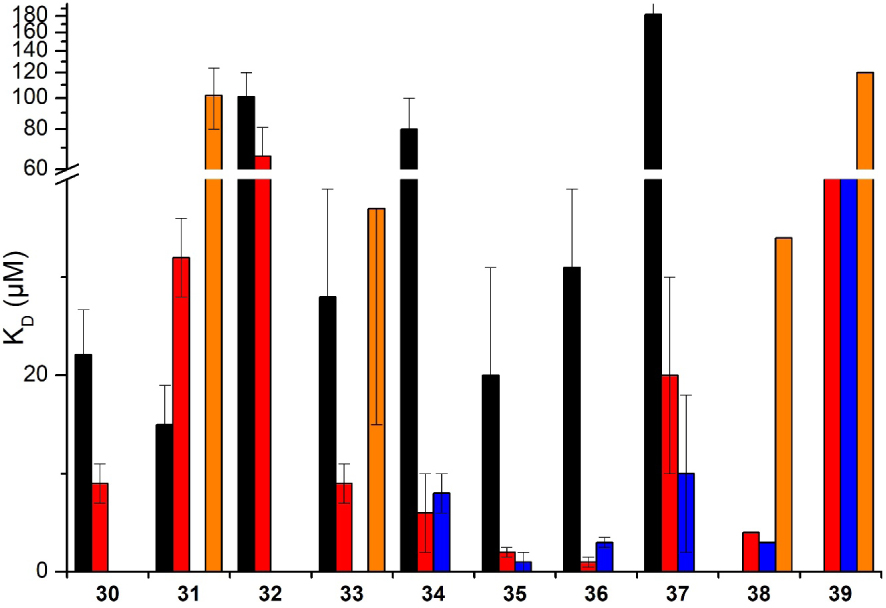
KD values obtained with different ruthenium and iridium derivatives 30–39 in interaction with G-quadruplexes 1 (black), 3 (blue) and 6 (red) and duplex DNA (orange). No histogram for duplex DNA means that the KD value could not be determined due to too weak interaction. The reported values are the means of representative independent experiments, and the errors provided are standard deviations from the mean. Each experiment was repeated at least three times.
A first series based on dph (dph = dipyrazino[2,3-a:2’,3′-h]phenazine) and bpph (bpph = benzo[a]pyrazino[2,3-h]phenazine) ancillary ligands (complexes 30–33) were studied using G-quadruplex systems 1 and 6 [62, 63]. It was shown that the removal of two non-chelating nitrogen atoms from the dph ligand in complex 30 to form the bpph ligand in complex 31 led to a huge impact on the interactions with G4 and duplex DNA. Indeed, a decrease of the affinity toward G4 structure 6 was observed (KD = 9 μM and 32 μM for complexes 30 and 31, respectively), while a weak affinity for duplex DNA could be measured with complex 31 (KD = 102 μM) thus leading to a partial loss of selectivity. The dph analogues therefore appeared to be slightly more selective toward G4 versus duplex DNA compared to their respective bpph analogues.
Another series was based on the CPIP (2-(4-chlorophenyl)-1H-imidazo[4,5-f][1,10]phenanthroline) and CPIPTAP ligands (complexes 34–37). Investigation by BLI indicated that these complexes displayed a good affinity for G-quadruplex DNA (KD around 1 and 3 μM for 36 and 35, respectively, for G4 system 3) and selectivity over duplex DNA. It was also noticed that their affinities were higher for G-quadruplex structures 3 and 6 which contain TTA loops, than for parallel-stranded quadruplex 1. This is consistent with interactions of the complexes with G-quadruplexes through mixed π-stacking with the guanine tetrad and further interactions with loops and grooves. Due to their photophysical properties, these complexes are able to react with DNA through type II photoreaction (i.e., formation of singlet oxygen) or through photo-induced charge transfer (PET). Interestingly, complexes 34 and 36 elicited a dramatic photo-cytotoxic effect, as 100% mortality was obtained upon irradiation of U2OS osteosarcoma cells in their presence, whereas very low mortality was observed in the dark at the same drug concentration [64].
The interaction of iridium complexes 38 and 39 with G-quadruplex DNA was also investigated [65]. KD values in the micromolar range was obtained for the G-quadruplex structures that fall within the range of those reported for similar ruthenium(II) complexes. However, a weak selectivity for the G-quadruplex structure versus duplex DNA was observed. This could be explained by the net positive charge of the IrIII complexes in comparison with the RuII complexes that could favor non-specific ionic interactions with DNA.
2.3.3. Study of the interaction with i-motif DNA
As described below, the stability of i-DNA strongly depends on pH. Conditions typically used for screening of ligand candidates with native i-DNA sequences employ low pH which, at the same time, leads to protonation of the ligands and favors their non-specific interactions with nucleic acids. In that context, we have recently designed constrained i-DNA5 which shows an increased stability and invariability of the i-motif structure in a broad pH range, allowing to use this scaffold as an i-DNA substrate at physiologically relevant conditions [33]. This constrained i-DNA 5 was thus used to investigate the interaction between previously reported i-motif DNA ligands (Figures 5 and 12) and folded or unfolded i-DNA in acidic (pH 5.5) and near-neutral (pH 6.5) conditions by using BLI.
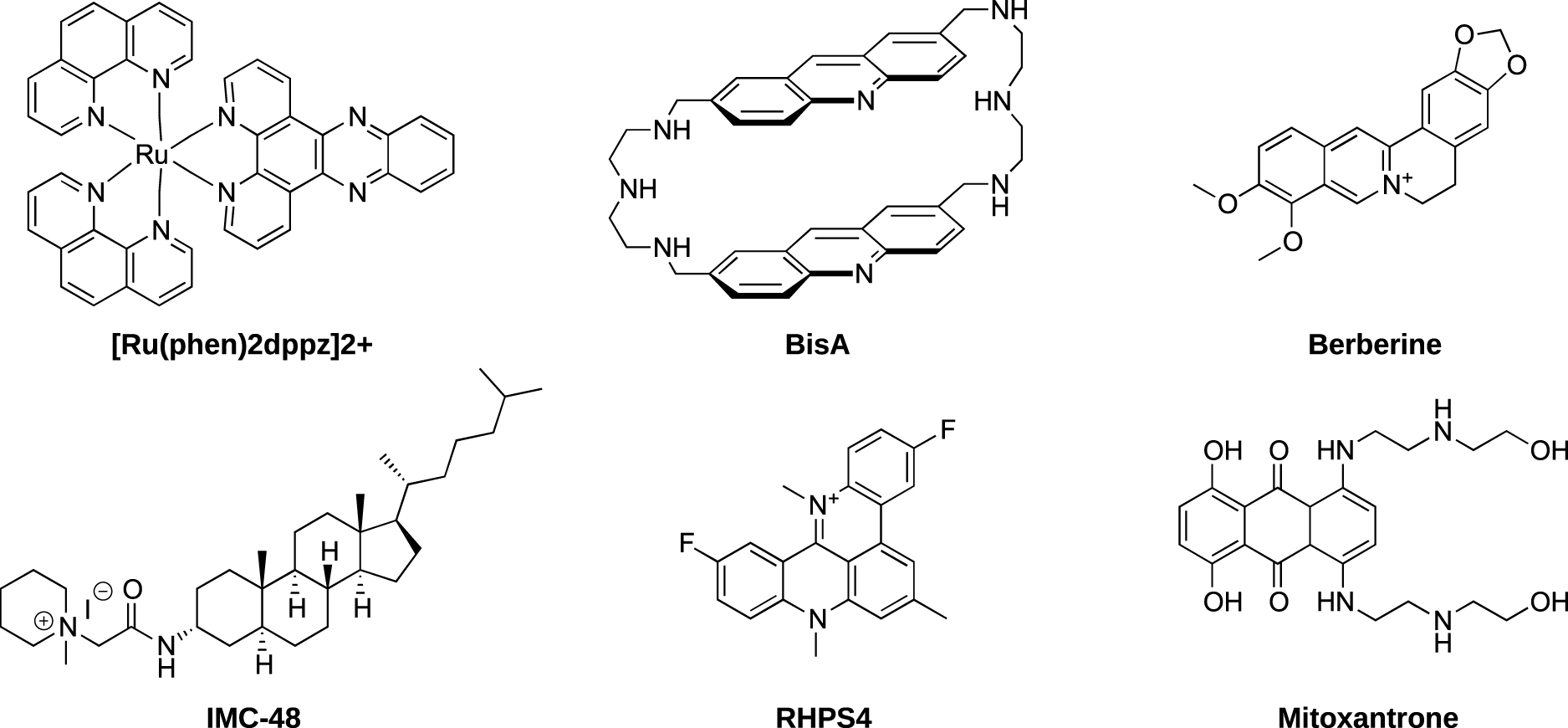
Structure of compounds used for the studies of i-motif interactions. [Ru(Phen)2dppz]2+ [66], BisA [67], Berberine [68], IMC-48 [19], RHPS4 [69] and mitoxantrone [70].
Very interestingly, we have observed that, despite several ligands such as macrocyclic bis-acridine (BisA) and pyridostatin (PDS) showing good affinities for the telomeric i-motif forming sequence, none of the ligands displayed selective interactions with the i-DNA structure nor were able to promote its formation (Figure 13) [71]. More recently we have reported that IMC-48, although described as i-DNA ligand, is a very weak ligand of i-DNA as no quantifiable interaction or significant stabilization of i-motif structures could be observed, stimulating a quest for an alternative mechanism of its biological activity [72]. All together, these results further emphasize the need for efforts to identify specific i-motif ligands. In this context, the use of constrained i-DNA5, which ensures an i-motif folding, represents an interesting alternative to identify unambiguous (i.e., affine and specific) i-DNA-interacting ligands.
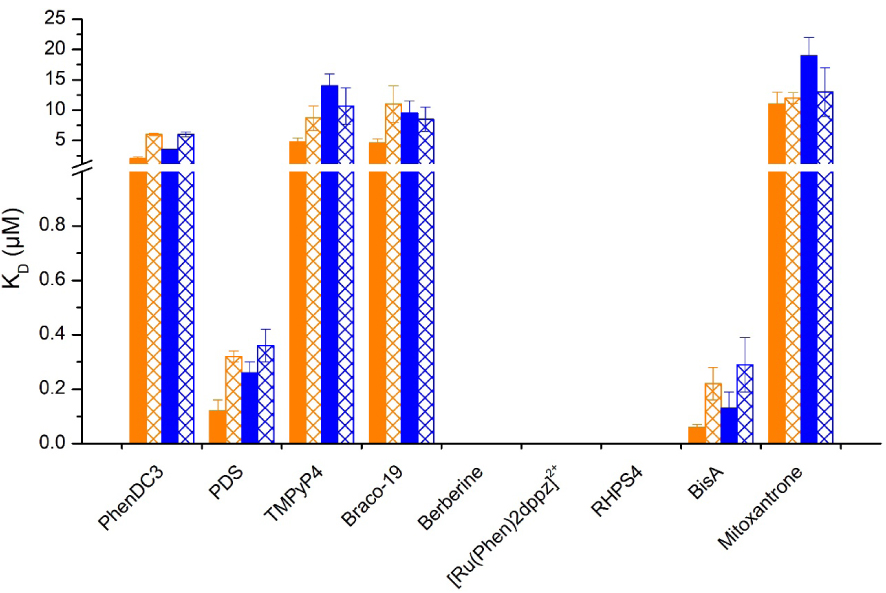
KD values obtained with different ligands in interaction with I-motif 6 (blue), h-Telo sequence (orange) at pH 5.5 and 6.5 fill and sparse pattern. No histogram for duplex berberine, [Ru(Phen)2dppz]2+ and RHPS4 means that the KD value could not be determined due to too weak interaction. The reported values are the means of representative independent experiments, and the errors provided are standard deviations from the mean. Each experiment was repeated at least three times.
2.4. Use of constrained G4 for protein fishing
Given the increasing roles of G4 structures in cellular metabolism, extensive researches have been conducted in the last years to identify new G4-dependent mechanisms. Notably classical pull-down approaches identified hundreds of proteins associated to G-rich oligonucleotides forming G4 structures [73]. However, in solution, G-rich single-stranded molecules are in equilibrium between unfolded and folded states, and thus numerous identified G4 binding proteins are also able to recognize unfolded G-rich sequences [74]. In this context, we have used the constrained G-quadruplex 3 which folds into the single antiparallel topology.
Moreover, such locked G4 displays a thermal stability significantly higher than unconstrained G4 which strongly reduces the possibility to form unfolded single-stranded sequences. We identified through affinity purifications coupled to mass spectrometry (MS)-based quantitative proteomics a set of human proteins associated to locked G4 structures. Notably, this approach allowed us to identify NELF proteins as a new G4-interacting complex, leading us to investigate the impact of RNA-Pol II pausing mechanism in the response to G4 stabilization by G4 ligands [75].
3. Conclusion
The different constrained G-quadruplex systems have proved efficient tools for the identification of G4 structure-specific synthetic ligands. They could also give some interesting information about the mode of interaction and the selectivity versus duplex and also versus G4 topologies. The latter could be crucial to design more specific G4 ligands associated with less off-target side effects. The constrained i-DNA represents also an interesting tool. Indeed, our recent study has demonstrated that all the molecules described so far as i-motif ligands, are not able to discriminate between folded and unfolded i-motif structures. The constrained i-DNA will thus be used to identify unambiguous (i.e., affine and specific) i-DNA-interacting ligands.
The pull-down strategy using constrained G-quadruplex DNA was proved efficient to identify proteins selective for a single G4 topology. Future directions of our approach will concern the construction of constrained G4 structures mimicking parallel G4 topologies in order to refine the impact of loops on protein binding. We will also use constrained i-DNA 5 to identify proteins interacting with the non-canonical secondary structure i-motif.
Lastly, given the fact that constrained G-quadruplexes reduce the unfolding to single-stranded sequences, we envisioned to use system 3 for the selection of topologically specific G4 antibody. In the same way, due to the lower sensitivity of constrained i-DNA to pH conditions, we will also use this system for the selection of specific i-motif antibodies.
Declaration of interests
The authors do not work for, advise, own shares in, or receive funds from any organization that could benefit from this article, and have declared no affiliations other than their research organizations.
Acknowledgements
The NanoBio-ICMG platforms (UAR 2607) are acknowledged for their support. We thank also our collaborators for providing the different samples of products: M. P. Teulade-Fichou and A. Grand from Institut Curie Orsay, B. Elias from Université catholique de Louvain, G. Pratviel from CNRS Toulouse, F. Thomas and O. Jarjayes from Université Grenoble-Alpes. The authors are also grateful to all the students who contributed to those works.





 CC-BY 4.0
CC-BY 4.0
Vous devez vous connecter pour continuer.
S'authentifier