1 Introduction
Exploration and investigation of new metal–organic frameworks has increased extensively since 1990s because of their potential for applications, such as porous metal–organic frameworks as promising hydrogen storage materials [1,2], gas/liquid separation [3,4] and catalysis ability [5]. Many specific functions of these compounds could not be substituted by traditional inorganic or organic materials. In general, the strategy to design and construct metal–organic networks of a variety of structures is to elaborately select different metals with preferred coordination geometries and ligands with different shape, size and rigidity. In this paper, we report on a new compound synthesized using two flexible ligands 1,2-bis(4-pyridyl)ethane (bpe) and 1,10-decanedicarboxylic acid (H2dcc).
Several chain-like alkyl dicarboxylate ligands, such as HOOC(CH2)nCOOH (N = 3–8) have been previously employed to build new metal–organic frameworks [6–9]. However, dicarboxylate ligands with N > 8 used to synthesize new structures are quite limited [10]. Herein, we report synthesis and structure analysis of a new copper coordination compound containing an unusually long alkyl dicarboxylate (ddc) and a flexible ligand (bpe).
2 Preparation
All chemicals were used as purchased. A 0.05 M acetonitrile solution of cupric nitrate, 0.1 M DMF solution of H2ddc, 0.025 M ethanol solution of bpe and 0.1 M DMF solution of triethylamine (tea) were pre-prepared. First, a buffer solution of 4 ml ethylene glycol was added into the bottom of U-type tube, then, solution was carefully added via disposable pipettes into both arms concurrently. The left arm the U-type was added 6 ml of acetonitrile cupric nitrate; the right arm was added 2 ml of H2ddc/DMF, then 2 ml tea/DMF followed by 2 ml bpe/ethanol solution. After several weeks without disturbance, blue block-like crystals suitable for X-ray diffraction analysis were collected.
3 Structural characterization
Data collection on a selected single crystal was carried out at 298 K on an automated Enraf-Nonius CAD4 diffractometer equipped with graphite monochromatic Mo Kα radiation (λ, 0.71073 Å). The cell parameters were obtained from least-squares analyses of 25 computer-centered reflections within a range of 4.86° = θ = 11.21°. The data collections were monitored by three standard reflections every four hours. No decay was observed except the statistic fluctuation. Data collections and reduction were controlled by the CAD4/PC [11], and XCAD4-PC [12], respectively. At the end of the systematic data collection the raw intensities were corrected for Lorentz and polarization effects. Direct phase determination and subsequent difference Fourier map synthesis yielded the positions of all non-hydrogen atoms, which were subjected to anisotropic refinement. Hydrogen atoms were added geometrically with their thermal parameters set to 1.2 × Ueq of the parent non-hydrogen atoms. The final full-matrix, least-squares refinement on F2 was applied for all observed reflections [I >2 σ(I)]. All calculations were performed by using the SHELX97 software package [13]. Analytic expressions of atomic scattering factors were employed, and anomalous dispersion corrections were incorporated [14]. Crystal data and details of relevant parameters adopted in the data collection as well as crystal structure refinement are listed in Table 1. Additional materials have been deposited at the Cambridge Crystallographic Data Center, 12 Union Road, Cambridge CB21EZ, UK, as a supplementary publication No. 236288 available on request from the CCDC.
Crystallographic data of Cu(bpe)(ddc)
| Sample code | BF1-47A |
| Molecular formula | C24H32CuN2O4 |
| Molecular weight | 476.06 |
| Color and habit | Transparent blue, block-like |
| Crystal size | 0.16 × 0.09 × 0.04 mm3 |
| Crystal system | Triclinic |
| Space group | P (No. 2) |
| Unit cell parameters | a = 7.944(3) Å, α = 86.72(2)° |
| b = 8.714(3) Å, β = 86.72(2)° | |
| c = 9.366(3) Å, γ = 86.72(2)° | |
| V = 560.5(3) Å3, Z = 1 | |
| F(000) | 251 |
| Density (calc.) | 1.400 g cm–3 |
| Diffractometer | Enraf-Nonius CAD4 |
| Radiation | Graphite-monochromatized Mo Kα λ = 0.71073 Å |
| Refs. used for cell measurement | 25, 4.86° = θ = 11.21° |
| Standard reflections | (–1, 1, –2); (1, 1, –4); (0, 0, –4) |
| Intensity variation | ± 2.8% |
| Absorption coefficient | 0.1007 mm–1 |
| Transmission factor | 0.879 –1.000 |
| Scan type and rate | ω scan |
| Scan range | (0.65 + 0.35 tan θ)° |
| Data collection range | 0 = h = 9; –10 = k =10; –11 = l =11; 2.41°= θ = 25.97° |
| Rint (from merging of eqiv. refls.) | None |
| Reflections measured | |
| Total | 2203 |
| Unique (n): | 2203 |
| Observed [I ≥ 2σ(I)]: | 1522 |
| Number of variables, p | 146 |
| Extinction coefficient | None |
| Weighting scheme | W = 1/σ2 [Fo2 + (0.0482 P)2 + 0.1537 P], P = (Fo2 + 2 Fc2)/3 |
| R1a | 0.1109 (all data) 0.0560 (observed data) |
| WR2b | 0.1275 (all data) 0.1108 (observed data) |
| Goof (S)c | 1.052 |
| Largest and mean Δ/σ | 0.000, 0.000 |
| Largest peak and hole | + 0.372 to –0.324 e Å–3 |
a .
b .
c .
4 Results
The crystals of the title compound C24H32N2CuO4 were grown in a slowly diffusing solution of bpe, H2ddc and tea into a methanol solution of Cu(NO3)2 through a ethylene glycol medium. C24H32N2CuO4 crystallizes in a triclinic space group P. The atomic coordination, selected bond lengths and bond angles are listed in Tables 1 and 2, respectively. Fig. 1 shows the coordination environment of Cu(II). It has a distorted octahedral geometry and is situated at the inversion center, where two carboxylates groups of dcc ligands in the equatorial plane bond to Cu(II) in a bidentate mode, the bpe ligands bind to Cu(II) at two axial positions through their pyridine nitrogen atoms. The oxygens of the carboxylate groups are asymmetrically coordinated to the Cu with a short bond distance of 1.928 Å (Cu–O1) and longer bond distance 2.655 Å (Cu–O2). Pyridine groups coming from bpe at the apical positions give an identical C–N distance of 2.015 Å. The bond angles of O–Cu–N and N–Cu–N are 91.23° and 180°, respectively, and the bite angles of the carboxylate at the copper metal is 54.52° (Table 3). All bond distances and angles around the Cu center are consistent with those reported for similar structures [15,16]. Each metal center is connected to four adjacent metal centers by two bpe and dcc ligands to generate a 2D structure. Although there are two conformations (anti and gauche) of bpe existing in various structures [17], only the anti structure has been found in the title compound. It is interesting to note that every chelating carboxylate group of the long alkyl chain is bent to coordinate the metal resulting in carbon backbone in dcc with torsion angles of C7–C8–C9–C10 (59.79°), and C8–C9–C10–C11 (58.72°), whereas the average torsion angle in the rest of alkyl chain is ca. 175°. Due to the bending manner of the flexible ddc ligands, the alternating Cu and dcc zigzag chains are formed along <111> axis. Due to such a bending, the shape of the resultant 2D structure is quite different from those of normal girds [18] or parquet (basket weave) pattern [19,20] (see Fig. 2). Within the 2D net, each rectangle grid is 52-membered ring deformed to two ‘fused’ rectangles. In this grid, the distances between two adjacent copper centers separated by bpe and ddc are 13.67 and 13.34 Å, respectively. The fact that long alkyl chain of ddc is bent significantly to comply with the crystal architecture is quite different from the structure arrangement of the dicarboxylate ligands reported previously [10]. The 2D net is compressed significantly leaving no space for guest molecules to enter. Fig. 3 shows the packing of 2D nets, with a separation of adjacent nets ca. 2.5 Å.
Atomic coordinates and thermal parameters for Cu(bpe)(ddc). Estimated standard deviations are given in parentheses
| Atoms | x(σ) | y(σ) | z(σ) | Ueq (σ) |
| Cu | 0 | 0 | 0 | 0.039(1) |
| O(1) | –0.662(4) | 0.1449(4) | –0.2229(3) | 0.044(1) |
| O(2) | 0.1665(4) | –0.0558(5) | –0.2348(4) | 0.066(1) |
| N | –0.1466(5) | –0.1582(5) | 0.0083(4) | 0.041(1) |
| C(1) | –0.0983(7) | –0.3322(6) | 0.1024(6) | 0.058(1) |
| C(2) | –0.2020(8) | –0.4388(7) | 0.1171(6) | 0.072(2) |
| C(3) | –0.3649(8) | –0.3700(7) | 0.0366(6) | 0.068(2) |
| C(4) | –0.4118(6) | –0.1926(7) | –0.0632(6) | 0.059(1) |
| C(5) | –0.2999(6) | –0.0923(6) | –0.0751(5) | 0.048(1) |
| aC(6 A) | –0.5017(12) | –0.4727(11) | 0.0652(9) | 0.066(2) |
| bC(6B) | –0.4130(40) | –0.5210(30) | 0.0160(30) | 0.066(2) |
| C(7) | 0.0493(6) | 0.0846(6) | –0.2987(6) | 0.047(1) |
| C(8) | 0.0349(6) | 0.1976(6) | –0.4769(5) | 0.053(1) |
| C(9) | 0.1979(6) | 0.1681(7) | –0.5614(6) | 0.058(1) |
| C(10) | 0.3569(6) | 0.1993(6) | –0.5098(6) | 0.061(1) |
| C(11) | 0.3350(6) | 0.3819(6) | –0.5225(6) | 0.060(1) |
| C(12) | 0.5060(6) | 0.4082(6) | –0.4873(6) | 0.061(1) |
| H(1 A) | 0.0102 | –0.3826 | 0.1602 | 0.069 |
| H(2 A) | –0.1619 | –0.5596 | 0.1824 | 0.086 |
| H(4 A) | –0.5191 | –0.1397 | –0.1229 | 0.071 |
| H(5 A) | –0.3338 | 0.0274 | –0.1451 | 0.057 |
| aH(6AA) | –0.4736 | –0.5763 | 0.1666 | 0.079 |
| aH(6AB) | –0.6177 | –0.3982 | 0.0662 | 0.079 |
| bH(6BA) | –0.3511 | –0.5399 | –0.0703 | 0.079 |
| bH(6BB) | –0.3757 | –0.6304 | 0.1120 | 0.079 |
| H(8 A) | –0.0570 | 0.1766 | –0.5239 | 0.063 |
| H(8B) | –0.0024 | 0.3212 | –0.4962 | 0.063 |
| H(9 A) | 0.1697 | 0.2449 | –0.6737 | 0.069 |
| H(9B) | 0.2299 | 0.0468 | –0.5489 | 0.069 |
| H(10 A) | 0.3920 | 0.1148 | –0.4003 | 0.073 |
| H(10B) | 0.4525 | 0.1741 | –0.5729 | 0.073 |
| H(11 A) | 0.2508 | 0.4027 | –0.4489 | 0.072 |
| H(11B) | 0.2875 | 0.4687 | –0.6287 | 0.072 |
| H(12 A) | 0.5475 | 0.3282 | –0.3778 | 0.074 |
| H(12B) | 0.5931 | 0.3756 | –0.5538 | 0.074 |
a Ueq. defined as one third of the trace of the orthogonalized U tensor.
b The S.O.F.s for a and b are 0.765 and 0.235, respectively.

ORTEP (50% probability) drawing and atom labeling diagram for Cu(bpe)(ddc).
Selected bond distances (Å) and bond angles (°) for Cu(bpe)(ddc)
| Cu–O(1)#1 | 1.928(3) | C(3)–C(4) | 1.371(7) |
| Cu–O(1) | 1.928(3) | C(3)–C(6 A) | 1.536(7) |
| Cu–N#1 | 2.015(3) | C(3)–C(6B) | 1.557(16) |
| Cu–N | 2.015(3) | C(4)–C(5) | 1.381(6) |
| Cu–O(2) | 2.655(4) | C(6 A)–C(6 A)#2 | 1.486(17) |
| Cu–O(2)#1 | 2.655(4) | C(6B)–C(6B)#2 | 1.35(6) |
| O(1)–C(7) | 1.265(5) | C(7)–C(8) | 1.521(6) |
| O(2)–C(7) | 1.232(5) | C(8)–C(9) | 1.497(6) |
| N–C(5) | 1.324(5) | C(9)–C(10) | 1.510(6) |
| N–C(1) | 1.337(5) | C(10)–C(11) | 1.504(6) |
| C(1)–C(2) | 1.367(6) | C(11)–C(12) | 1.516(6) |
| C(2)–C(3) | 1.375(7) | C(12)–C(12)#3 | 1.487(9) |
| O(1)#1–Cu–O(1) | 180.0(3) | N–C(1)–C(2) | 122.5(5) |
| O(1)#1–Cu–N#1 | 91.23(12) | C(1)–C(2)–C(3) | 120.9(5) |
| O(1)–Cu–N#1 | 88.77(12) | C(4)–C(3)–C(2) | 116.3(4) |
| O(1)#1–Cu–N | 88.77(12) | C(4)–C(3)–C(6 A) | 118.8(6) |
| O(1)–Cu–N | 91.23(12) | C(2)–C(3)–C(6 A) | 124.5(5) |
| N#1–Cu–N | 180.0(2) | C(4)–C(3)–C(6B) | 128.0(12) |
| O(1)#1–Cu–O(2) | 125.48(12) | C(2)–C(3)–C(6B) | 108.2(13) |
| O(1)–Cu–O(2) | 54.52(12) | C(6 A)–C(3)–C(6B) | 34.8(10) |
| N#1–Cu–O(2) | 90.48(12) | C(3)–C(4)–C(5) | 120.0(4) |
| N–Cu–O(2) | 89.52(12) | N–C(5)–C(4) | 123.2(4) |
| O(1)#1–Cu–O(2)#1 | 54.52(12) | C(6 A)#2–C(6 A)–C(3) | 107.8(8) |
| O(1)–Cu–O(2)#1 | 125.48(12) | C(6B)#2–C(6B)–C(3) | 110(3) |
| N#1–Cu–O(2)#1 | 89.52(12) | O(2)–C(7)–O(1) | 123.2(5) |
| N–Cu–O(2)#1 | 90.48(12) | O(2)–C(7)–C(8) | 121.4(5) |
| O(2)–Cu–O(2)#1 | 180.00(14) | O(1)–C(7)–C(8) | 115.4(4) |
| C(7)–O(1)–Cu | 107.6(3) | C(9)–C(8)–C(7) | 116.5(4) |
| C(7)–O(2)–Cu | 74.2(3) | C(8)–C(9)–C(10) | 116.9(4) |
| C(5)–N–C(1) | 116.9(4) | C(11)–C(10)–C(9) | 116.4(4) |
| C(5)–N–Cu | 121.1(3) | C(10)–C(11)–C(12) | 112.8(4) |
| C(1)–N–Cu | 122.0(3) | C(12)#3–C(12)–C(11) | 114.7(5) |
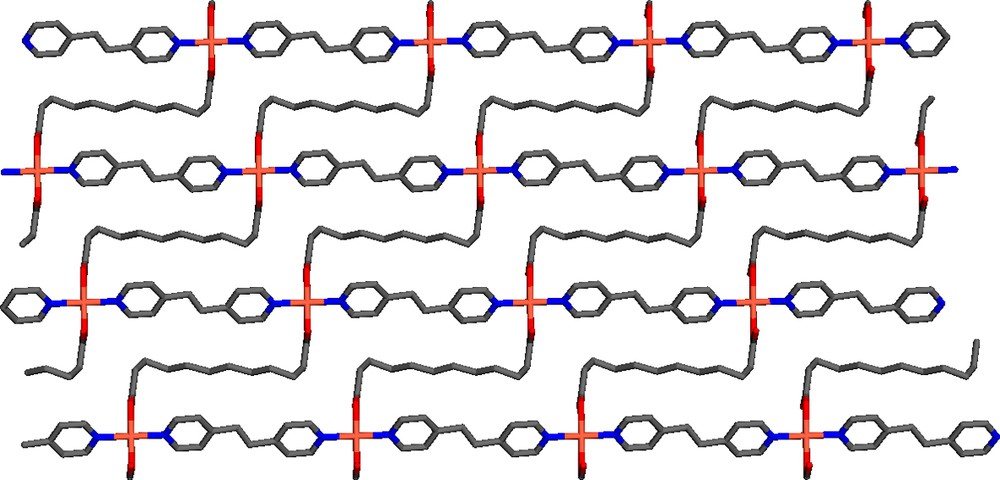
View of a single 2D net of Cu(bpe)(ddc) along a-axis.
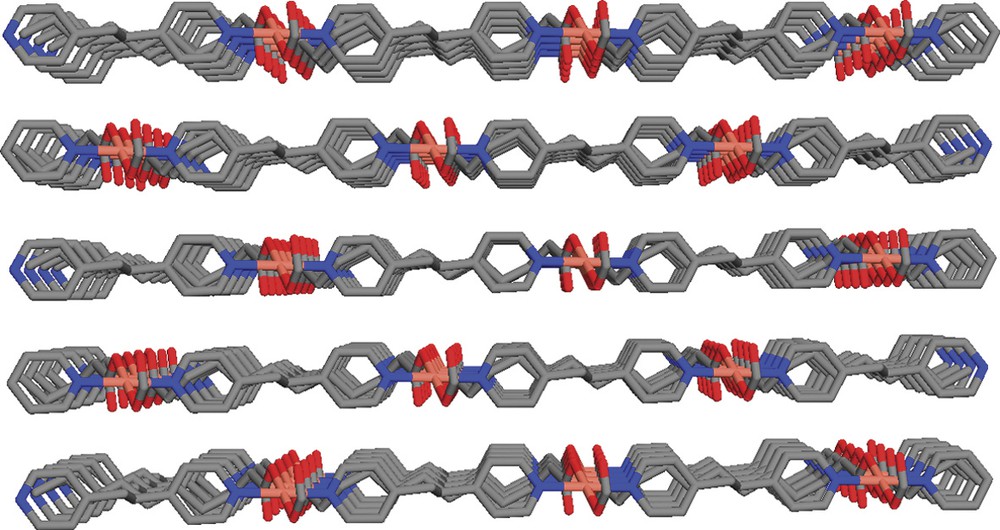
Perspective view of the crystal structure of Cu(bpe)(ddc) showing the packing of 2D nets.
5 Conclusion
In this paper, we describe the crystal growth of a new Cu-containing coordination polymer using diffusion method. We show that the crystal structure of Cu(bpe)(ddc) has an unusual shape due to the incorporation of a lengthy and flexible ligand, 1,10-H2dcc. The new structure consists of distorted octahedral of copper metal bound to two bpe as anti mode to form a linear chain propagating approximately along the b axis. The other coordination positions are chelated by ddc. The resultant structure is a 2D net. It is interesting to note each grid comprises of two fused rectangle-shipped rings as a result of ddc bending.
Acknowledgements
We gratefully acknowledge support from the National Science Foundation (DMR-0094872 and 0422932) and partial support by Rutgers University.


