1 Introduction
Of all the factors that affect agricultural productivity, the most powerful, and at the same time least modifiable, is the climate [1]. The phenotypic expression of a crop is the reflection of combined effect of genotype and environment. The phenotypic responses to changes in environment are not the same for all genotypes and the consequences of variation in genotypes are dependent on environment. This inter-play in effect of genetic and non-genetic components on phenotypic expression of a genotype is what we call genotype–environment interaction [2]. Better understanding of genotype–environment interaction is a basis for determining crop breeding strategies and provides useful information to identify stable genotypes over a range of environments [3]. Therefore, determination of the nature of genotype and environmental variations present in the plant characters and its magnitude are essential.
The geographical distribution of rice growing areas in different parts of the world reveals that rice is cultivated in the most diverse conditions, from 50° N to 35° S [4]. Adaptability of the rice plant to the environment is determined by its morphology and metabolic activity, which may vary according to the variety and growth stage. Differences in the metabolic pattern insure the pliability of adaptation and are reflected ultimately in the differences in morphological appearance of the plant as a whole [5]. Two types of adaptations are recognized. Agronomically, the wide adaptability of a rice variety refers to its high grain yield performance over diverse climatic conditions [6,7]. Specific adaptability is the ability of the rice plant to adjust to a specific adverse environmental condition e.g. deep water, salinity, drought, cold, etc.
Flowering response of a rice plant is the key indicator of crop duration. The subsequent ripening phase is thought to be of comparatively uniform extent of around 30 days [8]. Crop duration of rice cultivars determine their yield potential, local agronomic suitability and ability to escape from drought and natural hazards [9,10]. In irrigated systems, crop duration determines the calendar options for multiple rice cropping and intensified crop rotation [11,12]. Food crisis/availability and seasonal labour use pattern are also considerable issues for crop duration and planting dates. However, crop duration is interactively determined by the genotype and the environment [13].
Aromatic rices, though constitute a small group of rice in the consideration of consumption; it is a special group of rice that is regarded as best in quality [14]. Bangladesh (22–27° N, 88–93° E) has a stock of above 7000 rice germplasm of which around 100 are of aromatic type [15,16]. Aromatic rices are normally transplanted in rainy season (July–August) in Bangladesh and most of them are popularly grown in specific location. In Boro season (November–May), rice plant receives more solar energy because of clear sunshine in longer growth duration, and rationalize a possibility of higher yield. The increasing temperature during ripening period in this season may hamper aroma in kernel. However, obviously genotypic responses in this concern will be different. Higher yield with even mild aroma in some cultivars in Boro season may open a new avenue for increased aromatic rice production. Many workers have performed stability analysis with local and high yielding varieties of rice. But
2 Materials and methods
2.1 Identity of the genotypes and general experimental details
The experiment was conducted in 2004–2005. A total of 40 rice germplasm composed of 32 local aromatic, five exotic and three non-aromatic rice varieties as standard checks, were selected for this research (Table 1). Among the three non-aromatic varieties, BR28 was a modern Boro, BR39 was a modern T. Aman variety and the third one, Nizersail was used as a standard photoperiod sensitive genotype [17]. Exotic genotypes were collected from Pakistan (Basmati PNR346), Nepal (Sarwati and Sugandha-1) and Iran (Khazar and Neimat). The rest of the rice genotypes represented their distribution throughout Bangladesh. Forty rice genotypes formed the treatment variables and were assigned randomly to each unit plot of
Stability and response parameters for crop duration (days to flowering) of 40 genotypes.
| Sl# | Genotype | Mean |
|
|
|
| V1 | Badsha bhog Tapl-63 | 108.3 | −0.18 | 1.00 | 5.55 |
| V2 | Baoi jhak | 105.2 | −3.28 | 0.95 | 11.78 |
| V3 | Basmati Tapl-90 | 104.3 | −4.20 | 1.00 | 7.14 |
| V4 | Basmati PNR 346 | 98.2 | −10.26 | 0.98 | 36.93∗ |
| V5 | Begun bichi | 104.7 | −3.78 | 0.91 | 11.25 |
| V6 | Benaful | 111.1 | 2.64 | 1.21∗ | 10.87 |
| V7 | Bhog ganjia | 106.6 | −1.86 | 1.02 | 3.81 |
| V8 | BRRIdhan28 | 96.2 | −12.32 | 0.94 | 45.97∗ |
| V9 | BRRIdhan38 | 110.2 | 1.72 | 0.84∗ | 14.64 |
| V10 | BRRIdhan39 | 102.7 | −5.80 | 0.93 | 12.03 |
| V11 | Chinigura | 108.2 | −0.26 | 0.99 | 12.08 |
| V12 | Chinikani | 110.6 | 2.09 | 0.80∗ | 9.02 |
| V13 | Darshal | 111.0 | 2.53 | 0.80∗ | 6.97 |
| V14 | Doiar guro | 111.2 | 2.68 | 0.80∗ | 6.23 |
| V15 | Elai | 110.1 | 1.61 | 1.02 | 7.72 |
| V16 | Gandho kasturi | 166.2 | 57.68 |
|
|
| V17 | Gandhoraj | 107.7 | −0.84 | 0.86∗ | 19.45 |
| V18 | Hatisail Tapl-101 | 107.9 | −0.61 | 0.92 | 17.04 |
| V19 | Jamai sohagi | 105.6 | −2.93 | 0.94 | 22.90 |
| V20 | Jata katari | 105.8 | −2.72 | 0.93 | 8.14 |
| V21 | Jesso balam Tapl-25 | 107.0 | −1.47 | 0.87∗ | 5.35 |
| V22 | Jira katari | 107.4 | −1.11 | 0.89∗ | 5.92 |
| V23 | Kalijira Tapl-73 | 113.5 | 5.01 | 0.78∗ | 24.77 |
| V24 | Kalomai | 106.3 | −2.22 | 0.92 | 10.37 |
| V25 | Kamini soru | 105.7 | −2.80 | 0.88∗ | 6.70 |
| V26 | Kataribhog | 105.4 | −3.14 | 0.91 | 9.26 |
| V27 | Khazar | 105.4 | −3.05 | 0.86∗ | 30.68∗ |
| V28 | Laljira Tapl-130 | 110.5 | 2.01 | 0.85∗ | 28.77 |
| V29 | Niemat | 102.0 | −6.51 | 0.95 |
|
| V30 | Nizersail | 109.9 | 1.39 | 1.08 | 24.07 |
| V31 | Philippine katari | 105.2 | −3.28 | 0.96 | 9.43 |
| V32 | Premful | 106.6 | −1.93 | 0.84∗ | 1.83 |
| V33 | Radhuni pagal Tapl-77 | 115.1 | 6.64 | 0.82∗ | 10.71 |
| V34 | Rajbhog | 110.6 | 2.14 | 0.85∗ | 12.51 |
| V35 | Sai bail | 106.9 | −1.59 | 0.94 | 7.59 |
| V36 | Sakkor khora | 108.8 | 0.28 | 0.86∗ | 8.02 |
| V37 | Sarwati | 104.5 | −4.01 | 0.93 | 35.08∗ |
| V38 | Sugandha-1 | 103.3 | −5.16 | 0.89∗ | 29.73∗ |
| V39 | Tilkapur | 108.4 | −0.14 | 0.88∗ | 17.54 |
| V40 | Ukni madhu | 105.4 | −3.11 | 0.91 | 11.30 |
2.2 Identity of the environments
The four locations were:
B = Benarpota Farm, BRRI Regional Station, Satkhira (22.72° N, 89.08° E).
C = Charchandia Farm, BRRI Regional Station, Sonagazi, Feni (22.84° N, 91.39° E).
D = Domar Seed Production Farm, BADC, Sonaroy, Domar, Nilphamari (26.10° N, 88.84° E).
H = Headquarter Farm, Bangladesh Rice Research Institute (BRRI), Gazipur (24.00° N, 90.42° E).
Seed sowing was done in four dates. Three dates were in T. Aman (with an interval of 20 days) and one date was in Boro season.
1 = 1st planting in T. Aman (sowing in seedbed on 16th July 2004).
2 = 2nd planting in T. Aman (sowing in seedbed on 5th August 2004).
3 = 3rd planting in T. Aman (sowing in seedbed on 25th August 2004).
4 = Planting in Boro season (sowing in seedbed on 5th November 2004).
The combination of two factors (locations × planting times) resulted a total of 16 environments viz. B1, B2, B3, B4, C1, C2, C3, C4, D1, D2, D3, D4, H1, H2, H3 and H4.
2.3 Statistical analysis
Stability analysis was done according to the regression model of Eberhart and Russel [19]. The stability parameters viz. phenotypic index (
3 Results and discussion
3.1 Significance of mean squares
There were high genetic variability among the genotypes. Differences among the environments were also highly pronounced and influence on environmental differences on crop duration was immense. The genotype × environment interactions were also highly significant (Table 2). Thus the data were extended for analysis of stability indices. Significant genetic and environmental variability and significant genotype × environment interactions were reported for different characters in rice by several workers [22,23].
AMMI4 analysis of variance for the days to flowering data of rice genotypes.
| Source | df | SS | MS | F |
| Total | 1919 | 484067 | 252.2 | – |
| Treatments | 639 | 483752 | 757.0 |
|
| Genotypes | 39 | 63009 | 1615.6 |
|
| Environments | 15 | 314629 | 20975.3 |
|
| G × E interactions | 585 | 106429 | 181.9 |
|
| IPCA1 | 53 | 98299 | 1854.7 |
|
| IPCA2 | 51 | 5067 | 99.4 |
|
| IPCA3 | 49 | 893 | 18.2 |
|
| IPCA4 | 47 | 512 | 10.9 |
|
| G × E residual | 385 | 1658 | 4.3 |
|
| Error | 1280 | 315 | 0.2 | – |
3.2 Stability and response parameters
The response and stability parameters along with mean performance and phenotypic index for crop duration (days to flowering) are presented in Table 1. The range of genotypic means over the environments was found between 96 to 166 days. The lowest crop duration was obtained for V8 (96 days) followed by V4 (98 days) and V29 (102 days). The environment means were in the range of 91–155 days (see Supplementary Material Appendix I).
Twenty-seven genotypes had negative phenotypic indices (
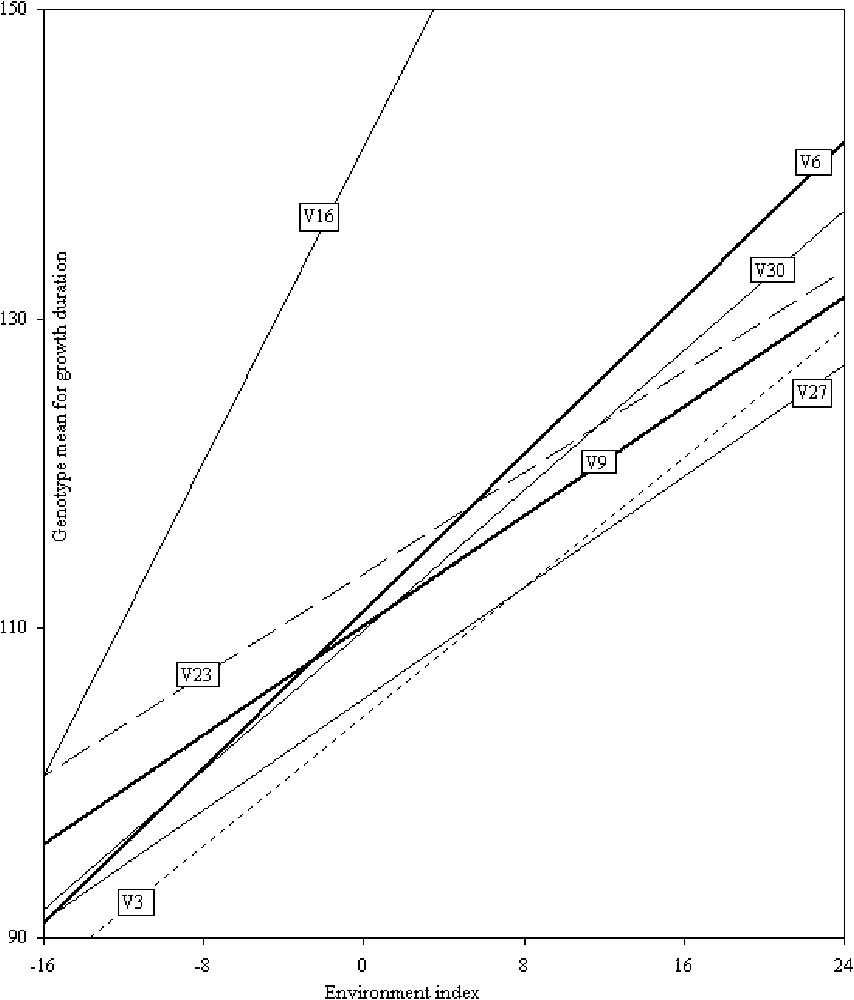
Linear regression showing the influence of different environments on the crop duration (days to flowering) of rice genotypes.
3.3 Measurement of interaction effects through AMMI model
The effect of genotype, environment and the components of
AMMI4 model for the crop duration (days to flowering) data; grand mean is 108.48 days.
| Genotypes/environments | Deviation (days) | IPCA score |
|||
| IPCA 1 | IPCA 2 | IPCA 3 | IPCA 4 | ||
| V1 | −0.18 | −0.11 | −0.57 | −0.15 | −0.89 |
| V2 | −3.28 | −0.38 | −1.09 | −0.17 | −0.84 |
| V3 | −4.20 | 0.03 | 0.42 | −1.18 | 0.58 |
| V4 | −10.26 | 0.14 | 2.60 | −0.64 | 0.01 |
| V5 | −3.78 | −0.54 | −1.15 | −0.23 | −0.76 |
| V6 | 2.64 | 0.99 | −1.28 | 0.65 | 0.25 |
| V7 | −1.86 | 0.13 | 0.21 | −0.40 | 0.04 |
| V8 | −12.32 | −0.04 | 2.95 | −0.76 | −0.16 |
| V9 | 1.72 | −0.88 | −0.64 | −0.15 | 1.67 |
| V10 | −5.80 | −0.19 | 1.42 | −0.22 | 0.15 |
| V11 | −0.26 | −0.15 | −0.95 | 0.89 | −1.23 |
| V12 | 2.09 | −1.08 | −0.22 | 0.56 | 1.03 |
| V13 | 2.53 | −1.08 | −0.19 | 0.72 | 0.68 |
| V14 | 2.68 | −1.05 | 0.27 | 0.39 | 0.28 |
| V15 | 1.61 | 0.16 | 0.72 | 1.06 | 0.09 |
| V16 | 57.68 | 17.26 | −0.38 | 0.12 | 0.23 |
| V17 | −0.84 | −0.87 | −1.31 | −0.51 | −1.27 |
| V18 | −0.61 | −0.51 | −1.50 | −0.12 | −0.69 |
| V19 | −2.93 | −0.25 | 1.24 | 2.46 | −0.15 |
| V20 | −2.72 | −0.40 | −0.75 | −0.50 | −0.89 |
| V21 | −1.47 | −0.68 | −0.36 | −0.35 | 0.18 |
| V22 | −1.11 | −0.55 | −0.33 | −0.45 | −0.19 |
| V23 | 5.01 | −1.30 | −0.90 | 1.06 | 1.85 |
| V24 | −2.22 | −0.53 | −1.13 | −0.27 | 0.12 |
| V25 | −2.80 | −0.59 | 0.25 | −1.43 | 0.53 |
| V26 | −3.14 | −0.43 | −0.45 | −1.29 | 0.63 |
| V27 | −3.05 | −0.53 | 2.02 | −1.57 | 0.12 |
| V28 | 2.01 | −0.92 | −1.34 | −0.17 | 1.55 |
| V29 | −6.51 | 0.06 | 3.58 | 0.28 | −0.20 |
| V30 | 1.39 | 0.20 | −1.97 | −1.05 | 0.34 |
| V31 | −3.28 | −0.25 | −0.84 | −1.15 | −0.90 |
| V32 | −1.93 | −0.81 | 0.02 | −0.05 | −0.50 |
| V33 | 6.64 | −0.99 | −0.50 | 0.59 | 0.45 |
| V34 | 2.14 | −0.89 | −0.86 | −0.11 | 0.21 |
| V35 | −1.59 | −0.38 | −0.10 | 1.28 | −0.36 |
| V36 | 0.28 | −0.81 | −0.50 | 0.43 | 0.64 |
| V37 | −4.01 | −0.10 | 2.62 | −0.27 | −0.17 |
| V38 | −5.16 | −0.34 | 2.40 | 0.86 | −0.42 |
| V39 | −0.14 | −0.76 | −0.43 | 1.99 | −0.62 |
| V40 | −3.11 | −0.55 | −0.96 | −0.13 | −1.44 |
| B1 | −4.62 | −2.24 | −3.66 | 0.14 | −0.60 |
| B2 | −17.50 | −2.68 | 0.09 | 0.49 | −0.41 |
| B3 | −16.60 | −2.75 | 1.55 | −0.30 | 0.01 |
| B4 | 16.23 | 7.81 | 0.26 | −0.55 | −1.00 |
| C1 | −4.61 | −2.46 | −3.31 | 0.92 | 0.91 |
| C2 | −16.60 | −2.74 | 0.26 | −0.48 | 1.25 |
| C3 | −13.10 | −2.64 | 2.25 | −1.32 | −0.70 |
| C4 | 12.31 | 7.64 | −0.14 | 0.26 | 0.72 |
| D1 | −2.14 | −2.46 | −3.38 | 0.85 | −1.54 |
| D2 | −13.50 | −2.63 | 0.56 | −1.49 | −2.78 |
| D3 | −11.00 | −2.18 | 3.93 | 4.33 | −0.31 |
| D4 | 28.64 | 7.22 | −0.28 | 0.23 | 0.32 |
| H1 | −4.71 | −2.34 | −2.04 | 0.07 | 2.15 |
| H2 | −16.60 | −2.67 | 0.96 | −1.14 | 0.30 |
| H3 | −15.40 | −2.86 | 2.60 | −1.81 | 1.65 |
| H4 | 12.40 | 7.96 | 0.36 | −0.19 | 0.03 |
Fig. 2 showed the expected crop duration considering mean duration on the abscissa and IPCA1 scores (for genotypes and environments) on the ordinate. Sixteen environments are shown with open circles. Filled tetragons denote the genotypes. The genotypes with similar positions were not shown in the graph. Eighteen genotypes out of 40 were presented in the AMMI biplot. Straight lines draw attention to the grand mean on the abscissa and to zero on the ordinate.

AMMI1 model for crop duration (days to flowering) data, accounting for 97.7% of the treatment SS.
The genotypes V6, V8 and V37 showed more or less similar distances from the horizontal reference line. However, series of displacements along the abscissa indicated their differences due to only main effects. On the other hand, V6, V9, V15, V28 and V30 are being dispersed along the ordinate and apparently they differ only in interaction effects. The genotypes V8 and V28 differ in both; while V1, V10, V20, V27 and V38 are rather similar with respect to both main effects and interaction effects.
The main effect for genotypes reflects breeding advances and the main effect for environments characterize the site [21]. Considering these responses, the four environments viz. B4, C4, D4 and H4 had exhibited remarkably longer life span of rice plants (Table 3 and Fig. 3). On the other hand, environments B2, C2, H2, B3 and C3 were characterized by extremely short duration. In general, shorter duration of rice varieties produces inferior yield mainly because of lower amount of solar radiation received.

AMMI2 model for the interaction of crop duration (days to flowering) data.
Direction and level of interactions of genotypes with environments could be determined from Fig. 2. For example, V16 had strong positive interactions with the environments B4, C4, D4 and H4, and strong negative interactions with all other 12 environments. In fact, this variety could not emerge panicle at all in the Boro season in all four locations. As a result, its duration was extended up to the next T. Aman crop. On the other hand, V9, V28 and V33 had strong positive interactions with D3, little interactions with B1 and H1, and strong negative interactions with B4, C4, D4 and H4. In general, local aromatic rice varieties possess photoperiod sensitivity at varying degrees. For this reason, life span might be flexible depending on planting time and season. Negative interaction for duration may be preferred up to a certain limit of grain yield reduction.
For further observation of interaction effects exclusively, a different type of biplot had been presented in Fig. 3. It held IPCA1 on the abscissa and IPCA2 on the ordinate. It captured 97% of the interaction as against 92% of Fig. 3. Figs. 2 and 3 together effectively captured 99% of the treatment SS in the AMMI2 model, leaving a RMS residual of only 1.26 day, or 1.16% of the grand mean (Table 2). Principles of biplot graph for IPCA1 and IPCA2 were described by Kempton where he proved efficiency of this type of graph in the explanation of interactions [26]. The genotypes V1, V3, V7, V9, V15, V20 and V33 and the environments B2, C2 and D2 are situated very near the origin. It indicated a little interaction for the entities (varieties and environments). Therefore the genotypes V1, V3, V7, V9, V15, V20 and V33 are stable over the environments and the environments B2, C2 and D2 are more stable environments for all the rice genotypes.
Note


