1 Introduction
Pangolins (Manidae) are scaly mammals, sole representatives of the order Pholidota. According to the morphological analysis of Gaudin et al. [1], which was based on 395 osteological characters, the eight extant species of pangolins (Pholidota, Manidae) can be classified into three genera: (1) Manis representing the four Asian species (M. pentadactyla – Chinese pangolin, M. javanica – Sunda pangolin, M. culionensis – Philippine pangolin, and M. crassicaudata – Indian pangolin); (2) Phataginus for the two species of African tree pangolins (P. tricuspis – White-bellied pangolin, and P. tetradactyla – Black-bellied pangolin); and (3) Smutsia representing the two species of African ground pangolins (S. gigantea – Giant pangolin, and S. temminckii – Temminck's pangolin). In addition, Gaudin et al. [1] suggested that the family Manidae can be divided into two subfamilies, the Asian subfamily Maninae containing only the single genus Manis, and the African subfamily Smutsiinae uniting the genera Smutsia and Phataginus. Although the two species of African tree pangolins are sometimes considered as monotypic genera, i.e., Phataginus tricuspis and Uromanis tetradactyla [2], we consider here only three genera of pangolins (Manis, Phataginus and Smutsia), as proposed by Gaudin et al. [1].
In this study, the complete mitochondrial genome was sequenced for the three genera of Pholidota, as represented by the species M. javanica, P. tricuspis and S. temminckii, and comparisons were performed with all mitochondrial data available in GenBank, including the genomes of M. pentadactyla [3] and P. tetradactyla [4], as well as the sequences from five markers, i.e., two protein-coding genes, subunit I of the cytochrome c oxidase (COI) and cytochrome b (Cytb), two rRNA genes (12S and 16S), and the control region (CR). Our three main objectives were (1) to better understand the molecular evolution of the mitochondrial genome in the order Pholidota, (2) to examine phylogenetic relationships among pangolin species, and (3) to identify potential taxonomic inconsistencies at the species level.
2 Materials and methods
2.1 DNA extraction, amplification, and sequencing
Total DNA was extracted from tissue culture cells of M. javanica (Thailand, tissue code: 1999.508) and muscle samples of P. tricuspis (Gabon, tissue code: 2005.GLC14) and S. temminckii (Tanzania, tissue code: SUN106) using the QIAGEN DNeasy Tissue Kit (Qiagen, Hilden, Germany).
Twenty-five overlapping fragments of the mitochondrial genome were amplified by PCR using primers published in previous studies [5–8], as well as four new primers specially designed for this study: DLU400 or DLU400M1/12SL41 or 12SL200; 12SU1230/12SL2226M1LA; 12SU829/16SL518; 16SU365/16SL1056; 16SU946/N1L64; Uleu or U16S1421/LMet or LMet2; N1U840/N2L492; N2U354M2 or IleU/AsnL; TrpU/C1L705; UTyr or C1U246/C1L1017; C1U897M1/C2L15M1; SerU or SerUM1/A8L1; C2U603M4/C3L168; A6U654M1 or A6U654M2/GlyLM1; C3MANIU (5′- CAA TAT ATC AAT GAT GAC GTG A-3′)/LARGMANI (5′- GTT GAY TTG TTT GTG ATG CTC A-3′); C3U780 or C3U780M3/N4L366 or N4L366M2; UArg or UArgM1/N4L918M1 or N4L1071R; N4U681/Leu2L or Leu2LM1; Ser2U/N5L652 or N5L652M1; N5U501/N5L1214; N5U1146M5/N5L154M5; N6UT298 (5′- GGC TCA ATC AAA CTA TAC TTC CT-3′)/CBLT298 (5′-AGT TTC ATC ATG CTG AGA TG-3′); N6RU102 or N6RU102M1/CBL402 or CBL402M1; GluMA/ProMA; CBU162 or U844/L482; and UThr13/L482. Amplifications were done in 20 μl using 3 μl of Buffer 10× with MgCl2, 2 μl of dNTP (6.6 mM), 0.12 μl of Taq DNA polymerase (2.5 U, Qiagen, Hilden, Germany) and 0.75 μl of the two primers at 10 μM. The standard PCR conditions were as follows: 4 min at 94 °C; 5 cycles of denaturation/annealing/extension with 45 s at 94 °C, 1 min at 60 °C and 1 min at 72 °C, followed by 30 cycles of 30 s at 94 °C, 45 s at 55 °C, and 1 min at 72 °C, followed by 10 min at 72 °C.
Both strands of PCR products were sequenced using Sanger sequencing on an ABI 3730 automated sequencer at the Centre national de séquençage (Genoscope) in Évry (France). The sequences were edited and assembled using Sequencher 5.1 (Gene Codes, Corp., Ann Arbor, MI, USA).
2.2 Phylogenetic analyses
The analyses based on complete mitochondrial genomes included five species of the order Pholidota: M. javanica, M. pentadactyla, P. tetradactyla, P. tricuspis, and S. temminckii. Four genera, representing different orders of Laurasiatheria, were used to root the Pholidota tree: Boselaphus (Cetartiodactyla), Canis (Carnivora), Ceratotherium (Perissodactyla), and Rhinolophus (Chiroptera). GenBank accession numbers of the DNA sequences are provided on the tree of Fig. 2. Complete mitochondrial genomes were aligned using MUSCLE [9] and then further adjusted by eye with Se-Al v2.0a11 [10]. All ambiguous regions, i.e., involving ambiguity in the position of gaps, were excluded from the analyses to avoid erroneous hypotheses of primary homology. For this reason, the control region was not included in the final alignment. The length of the reduced alignment is 14,926 nt (available upon request to AH). The phylogenetic tree was constructed using PAUP* version 4 [11] based on the Maximum Likelihood (ML) method and the GTR + I + G model selected by the Akaike information criterion under jModelTest 2 [12]. Bootstrap percentages (BP) were computed using 1000 replicates.
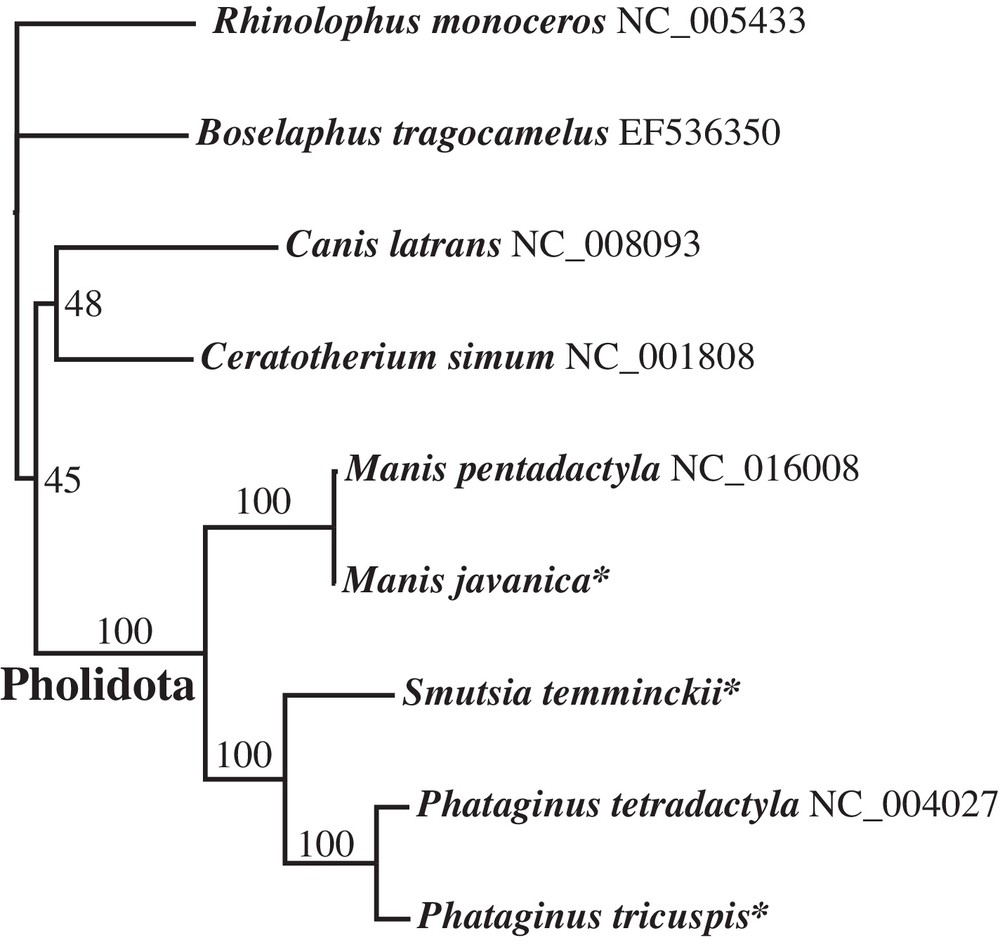
Interspecific relationships within the order Pholidota as inferred from a DNA alignment of complete mitochondrial genomes. Species labelled with an asterisk (*) were sequenced for this study. Values on the branches are bootstrap percentages B.
BLASTN searches were performed on NCBI [13] using the mtDNA genome of P. tricuspis as a query to extract all available mitochondrial sequences of Manidae. The sequences were filtered by mitochondrial markers (COI, Cytb, 12S, 16S, and CR) and aligned as explained above. Uncorrected nucleotide distances (D) were calculated in PAUP* for each datasets and the Neighbour-Joining (NJ) method was applied to reconstruct clusters of similar sequences, with BP computed after 1000 replicates.
3 Results and discussion
3.1 Characteristics of the mitochondrial genome
Protein-coding, ribosomal and transfer RNA genes were identified by comparison with the two published genomes of Pholidota, M. pentadactyla [3] and P. tetradactyla [4]. The annotated sequences of M. javanica, P. tricuspis, and S. temminckii were deposited in GenBank under accession numbers KP306514-KP306516. The three new mitochondrial genomes of Manidae represent circular double stranded DNA molecules that range between 16,559 and 16,576 bp in length, and that contains the 37 genes described in Table 1. Among these genes, only ND6 and eight tRNAs are encoded by the L-strand, whereas all the other genes are encoded by the H-strand. The gene arrangement is the same as that found in other mammalian genomes. All protein-coding genes of the mtDNA have a methionine start codon (ATR), except ND5 (ATT). Most protein-coding genes appear to be terminated by TAR, although this stop codon is incomplete in the COIII, ND3 and ND4 genes. In M. javanica, the COI, Cytb, and ND6 genes are terminated by AGA or AGG.
Characteristics of the mitochondrial genome in three species of the family Manidae.
| Name of gene | Location (Size in bp) | Strand | Start codon | Stop codon | ||
| M. javanica a | P. tricuspis b | S. temminckii c | ||||
| tRNA-Phe | 1–68 (68) | 1–68 (68) | 1–68 (68) | H | ||
| 12S ribosomal RNA (12S) | 69–1028 (960) | 69–1026 (958) | 69–1027 (959) | H | ||
| tRNA-Val | 1029–1094 (66) | 1027–1093 (67) | 1028–1093 (66) | H | ||
| 16S ribosomal RNA (16S) | 1095–2665 (1571) | 1094–2648 (1555) | 1094–2647 (1554) | H | ||
| tRNA-Leu (UUR) | 2666–2739 (74) | 2649–2723 (75) | 2648–2722 (75) | H | ||
| NADH dehydrogenase subunit 1 (ND1) | 2743–3699 (957) | 2727–3683 (957) | 2727–3683 (957) | H | ATG | TAA |
| tRNA-Ile | 3699–3767 (69) | 3683–3751 (69) | 3683–3751 (69) | H | ||
| tRNA-Gln | 3765–3837 (73) | 3749–3820 (72) | 3749–3820 (72) | L | ||
| tRNA-Met | 3839–3907 (69) | 3822–3890 (69) | 3822–3890 (69) | H | ||
| NADH dehydrogenase subunit 2 (ND2) | 3908–4948 (1041) | 3891–4934 (1044) | 3891–4934 (1044) | H | ATA | TAG |
| tRNA-Trp | 4947–5013 (67) | 4933–4998 (66) | 4933–5000 (68) | H | ||
| tRNA-Ala | 5017–5085 (69) | 5002–5070 (69) | 5003–5071 (69) | L | ||
| tRNA-Asn | 5087–5159 (73) | 5072–5144 (73) | 5073–5145 (73) | L | ||
| Origin of L-strand replication (OL) | 5159–5196 (38) | 5144–5180 (37) | 5145–5181 (37) | |||
| tRNA-Cys | 5193–5257 (65) | 5177–5241 (65) | 5178–5242 (65) | L | ||
| tRNA-Tyr | 5258–5324 (67) | 5242–5308 (67) | 5243–5309 (67) | L | ||
| Cytochrome c oxidase subunit I (COI) | 5326–6876 (1551) | 5310–6863 (1554) | 5311–6864 (1554) | H | ATG | AGAa AGGb,c |
| tRNA-Ser (UCN) | 6872–6940 (69) | 6855–6923 (69) | 6856–6924 (69) | L | ||
| tRNA-Asp | 6948–7014 (67) | 6931–6997 (67) | 6932–6998 (67) | H | ||
| Cytochrome c oxidase subunit II (COII) | 7015–7698 (684) | 6998–7681 (684) | 6999–7682 (684) | H | ATG | TAA |
| tRNA-Lys | 7701–7764 (64) | 7685–7750 (66) | 7685–7751 (67) | H | ||
| ATP synthase F0 subunit 8 (ATP8) | 7766–7969 (204) | 7752–7958 (207) | 7754–7954 (201) | H | ATG | TAAa,c TAGb |
| ATP synthase F0 subunit 6 (ATP6) | 7927–8606 (681) | 7913–8592 (681) | 7915–8594 (681) | H | ATG | TAA |
| Cytochrome c oxidase subunit III (COIII) | 8607–9391 (785) | 8593–9377 (785) | 8595–9379 (785) | H | ATG | TAN* |
| tRNA-Gly | 9391–9459 (69) | 9377–9444 (68) | 9379–9447 (69) | H | ||
| NADH dehydrogenase subunit 3 (ND3) | 9460–9806 (347) | 9445–9791 (347) | 9448–9794 (347) | H | ATA | TAN* |
| tRNA-Arg | 9807–9873 (67) | 9792–9858 (67) | 9795–9861 (67) | H | ||
| NADH dehydrogenase subunit 4L (ND4L) | 9874–10,170 (297) | 9859–10,155 (297) | 9862–10,158 (297) | H | ATG | TAA |
| NADH dehydrogenase subunit 4 (ND4) | 10,164–11,541 (1378) | 10,149–11,526 (1378) | 10,152–11,529 (1378) | H | ATG | TNN* |
| tRNA-His | 11,542–11,609 (68) | 11527–11594 (68) | 11530–11598 (69) | H | ||
| tRNA-Ser (AGY) | 11610–11668 (59) | 11595–11653 (59) | 11599–11657 (59) | H | ||
| tRNA-Leu (CUN) | 11670–11740 (71) | 11654–11723 (70) | 11657–11725 (69) | H | ||
| NADH dehydrogenase subunit 5 (ND5) | 11741–13561 (1821) | 11724–13544 (1821) | 11726–13546 (1821) | H | ATT | TAA |
| NADH dehydrogenase subunit 6 (ND6) | 13545–14069 (525) | 13528–14055 (528) | 13530–14057 (528) | L | ATG | AGAa TAGb,c |
| tRNA-Glu | 14070–14138 (69) | 14056–14124 (69) | 14058–14126 (69) | L | ||
| Cytochrome b (Cytb) | 14142–15281 (1140) | 14128–15267 (1140) | 14130–15269 (1140) | H | ATG | AGA |
| tRNA-Thr | 15282–15348 (67) | 15268–15336 (69) | 15270–15338 (69) | H | ||
| tRNA-Pro | 15348–15413 (66) | 15336–15401 (66) | 15338–15403 (66) | L | ||
| D-loop | 15414–16576 (1163) | 15402–16570 (1169) | 15404–16559 (1156) |
a Manis javanica.
b Phataginus tricuspis.
c Smutsia temminckii.
The control region is highly conserved among the five species of Manidae, both in structure and length (sequence length varies between 1156 and 1169 bp), with three tandem RS2 repeats of 78/79 bp in the L domain, which are located just after a partial RS2 motif, i.e., “YATGTATAATCGTGCAT” (pos. 15,453 in M. javanica).
The overall base composition of the mt genome showed marked differences between the two Asian species (Manis) and the three African species (Phataginus and Smutsia), with higher percentages of C (29.1 vs. 23.2–24.0%) and G nucleotides (14.7 vs. 13.3–13.8%), and lower percentages of A (33.1 vs. 33.2–33.9%) and T nucleotides (23.1 vs. 28.7–29.9%) (Fig. 1).

(Colour online.) Overall base composition on the L-strand of the mitochondrial genome of five species of Manidae. The overall base composition of M. javanica was found to be identical to that of M. pentadactyla. The tree below was reconstructed using the NJ clustering method and a matrix of pairwise distances calculated by summing the percentage differences obtained for each of the four nucleotides A, G, C and T.
3.2 Phylogeny and taxonomy
Maximum Likelihood analyses based on an alignment of 14,926 bp resulted in a robust tree (Fig. 2) with maximal BP (100%) for interspecific relationships within the family Manidae. The results are in full agreement with the morphological study of Gaudin et al. [1]: the genera Manis and Phataginus are found to be monophyletic and the family Manidae is divided into two geographic groups corresponding to the Asian subfamily Maninae (Manis) and the African subfamily Smutsiinae (Smutsia and Phataginus). However, the terminal branch lengths of the two species of Manis are unexpectedly short in the ML tree (Fig. 2). Nucleotide distances indicate that the mitochondrial genome of M. javanica is 99% identical to that of M. pentadactyla published by Qin et al. [3] (NC_016008), suggesting species misidentification for one of them. To solve this issue, the mitochondrial genomes were compared to all homologous fragments available for Pholidota in GenBank. Five mtDNA alignments were analysed using the NJ clustering method, i.e., COI (38 taxa and 658 nt), Cytb (35 taxa and 402 nt), 12S (38 taxa and 395 nt), 16S (25 taxa and 521 nt), and CR (56 taxa and 1264 nt). In Fig. 3, the COI tree shows the existence of six robust clusters (BP = 100) corresponding to the following species: M. javanica, M. pentadactyla, P. tricuspis, P. tetradactyla, S. gigantea and S. temminckii. On the one hand, the NC_016008 genome of M. pentadactyla fell into the M. javanica cluster (0.2 < D < 3.5 %), which is composed of representatives from China, Malaysia and Thailand. On the other hand, the NC_016008 genome appears to be highly divergent from COI sequences of M. pentadactyla from Taiwan and China (provinces of GuangDong, Hainan, and Hunan) (13.2 < D < 13.7 %). Similar results were found for three other markers (Cytb, 12S and CR) using mitochondrial sequences produced by independent teams (Appendices 1, 2 and 4). The analyses suggest, therefore, that the NC_016008 genome published in Qin et al. [3] belongs to M. javanica rather than M. pentadactyla. However, the specimen was collected from Guangxi, a Chinese province where M. javanica is not currently recorded (Fig. 4). Accordingly, two hypotheses can be proposed to explain the presence of M. javanica in Guangxi: (1) the specimen was not native to Guangxi, but was imported in China from a Southeast Asian country (Vietnam, Lao PDR, Myanmar, Cambodia, Malaysia, Thailand or Indonesia) to satisfy the persistent demand for meat and scales used in traditional medicines [14]; (2) alternatively, the species M. javanica may also occur in sympatry with M. pentadactyla in the Guangxi province, implying that the geographic range currently provided by the IUCN [2] for M. javanica is incomplete. To resolve this and to ensure that optimal conservation strategies are in place, the possible occurrence of M. javanica in Guangxi should be reassessed urgently.
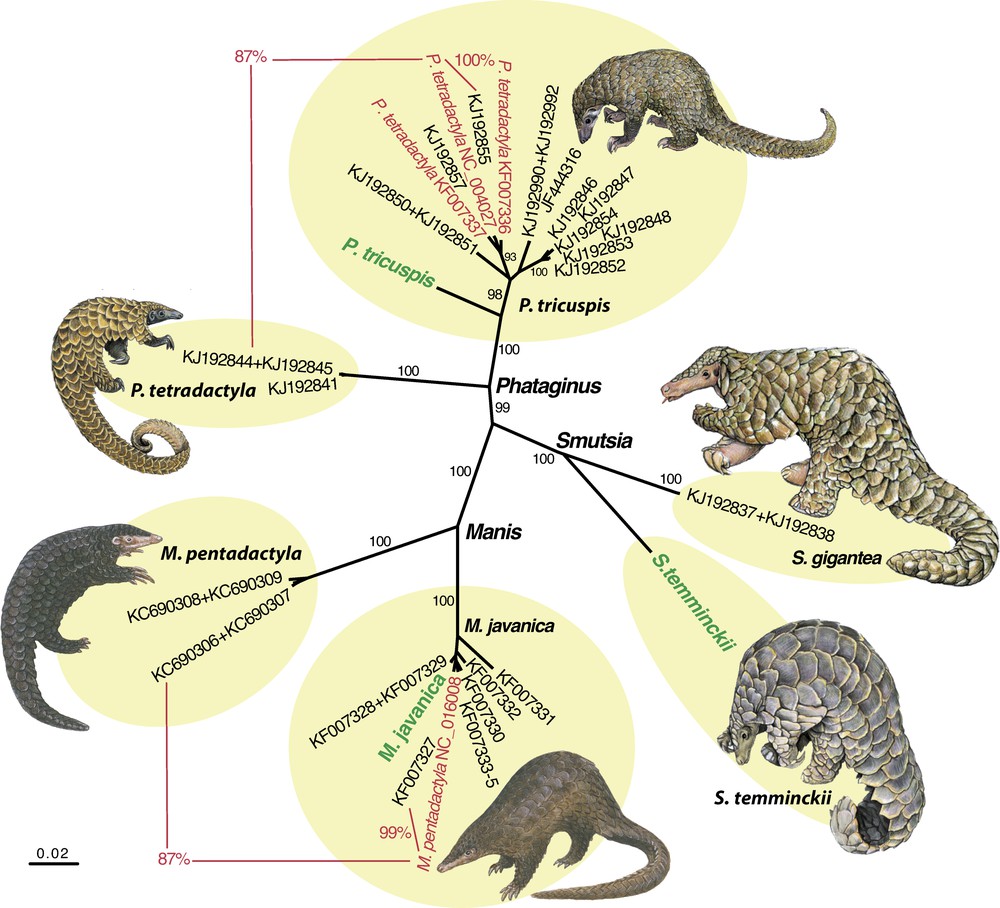
(Colour online.) Neighbour-joining tree reconstructed from COI sequences. Genomes sequenced for this study are indicated in green. Sequences from GenBank written in red are suspected to have been misidentified. The percentages in red represent nucleotide distances. Values on the branches are bootstrap percentages more than 90%. The six clusters highlighted in yellow correspond to the six following species of pangolin: Manis javanica, M. pentadactyla, Phataginus tricuspis, P. tetradactyla, Smutsia gigantea and S. temminckii. Illustrations are modified from Francis [18] for Manis species, and from Kingdon and Hoffmann [15] for Phataginus and Smutsia species.
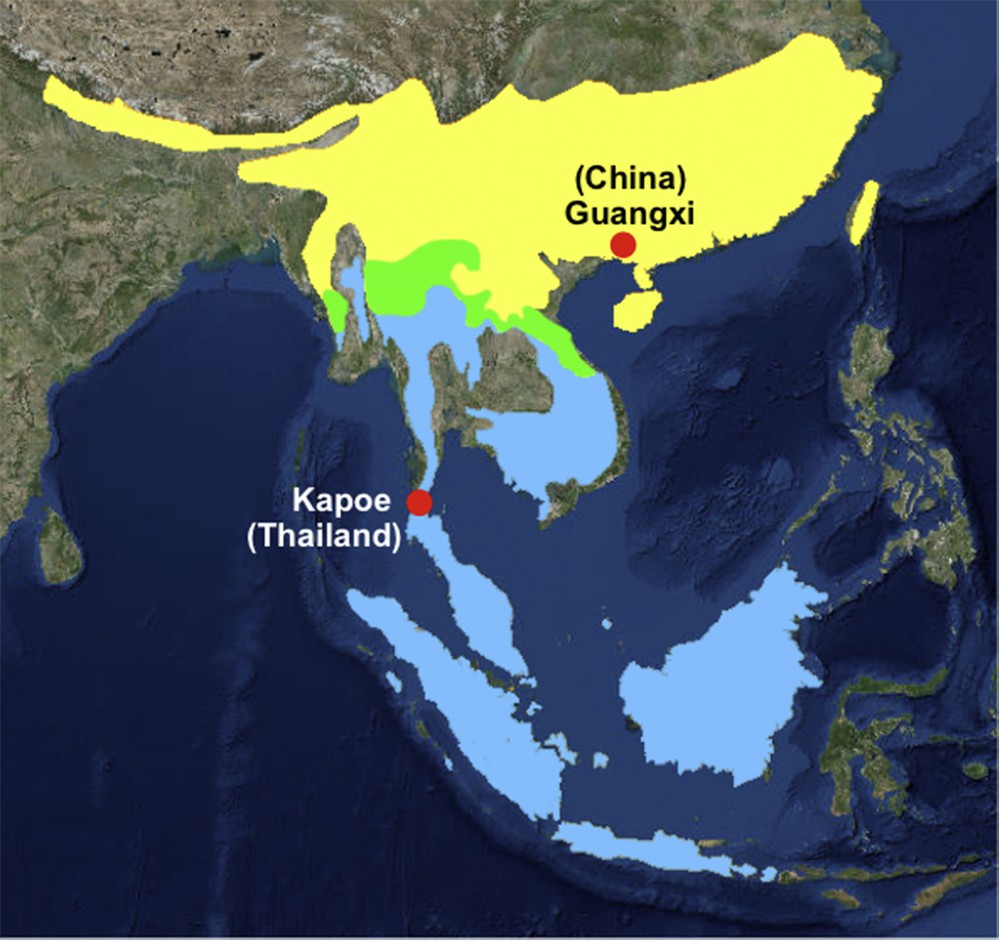
(Colour online.) Geographic distributions of Manis pentadactyla (in yellow) and M. javanica (in blue) (modified from the IUCN [2]). The overlapping ranges are represented in green. The complete mitochondrial genome of M. javanica was sequenced from an individual collected in Kapoe district (Thailand), whereas the NC_016008 genome of M. pentadactyla published by Qin et al. [3] came from the Guangxi province of China.
A similar problem was found for the mitochondrial genome of P. tetradactyla published by Arnason et al. [4]. Indeed, the NC_004027 genome is highly divergent from mitochondrial sequences of P. tetradactyla produced in different labs (COI: 13%; Cytb: 15%; 12S and 16S: 6%), whereas it shares 99–100% of identity with COI, Cytb, 12S, and 16S sequences of P. tricuspis obtained by independent teams (Fig. 3 and Appendices 1–3). Here again, the most likely hypothesis involves species misidentification of the specimen sequenced by Arnason et al. [4].
Within P. tricuspis, the analyses of mitochondrial data revealed high nucleotide divergence between the Gabonese pangolin sequenced for this study and the specimens collected in Ghana, Nigeria and Cameroon: 5.9–6.7% for COI, 8.0–9.5% for Cytb, 1.5–2.6% for 12S, 2.4–3.3% for 16S (Fig. 3 and Appendices 1–3). Given that mitochondrial markers are available for all species of pangolin currently recognized in Africa, and that the holotype of P. tricuspis was described from Guinea in West Africa [15], these comparisons suggest the existence of a new species of tree pangolin in Gabon. This finding could be consistent with recent studies supporting the existence of a strong biogeographic barrier (named charnière climatique) between Cameroon and Gabon for both animals and plants endemic to tropical rainforests [16,17]. Further studies are needed to determine the morphology, geographic distribution and conservation status of this new taxon.
Acknowledgements
The authors are grateful to people from the CIRMF (Centre international de recherches médicales de Franceville) who organized the fieldwork in Gabon: Philippe Blot, Éric Leroy, Xavier Pourrut, and André Délicat. We also acknowledge Vitaly Volobouev who provided tissue culture cells of Sunda pangolin, and Lorenzo Vinciguerra for the sample of Temminck's pangolin. We also thank the two anonymous reviewers for their comments on the first version of the manuscript. This work was supported by the “ATM Emergence” and the network “Bibliothèque du Vivant” funded by the CNRS, the Muséum national d’histoire naturelle, the INRA and the CEA (Genoscope).


