1 Introduction
The sorghum (Sorghum bicolor L. Moench) is one of the major cultivated cereals in the world.
It has been domesticated for approximately 3000 years before J.-C. in the Ethiopian regions of Africa [1–3]. The cultivated sorghum is subdivided into five races (guinea, kafir, durra, bicolor, and caudatum) and ten inter-races hybrids [4,5]. The sorghum is largely cultivated in Africa, where it is mostly the first human food. In fact, this spring cereal is known for its ability to survive the severe environmental conditions [6] and for the nutritious value of its grains (71% of starch, 10% of proteins, and 3% of lipids), which is similar to the case of the other cereals, justifying its importance in the arid and semi-arid regions. However, the production rates in Africa still remain weak compared to the rates of improved lineages grown in the USA with high yields (3 to 5 t/ha), because cultivated sorghums in Africa consist of local varieties with low yields (0.5 to 0.7 t/ha) [7]. The improvement of the productivity of this species constitutes a priority to overcome the demographic explosion of the African population [7].
The sorghum was introduced into Morocco with Islamisation, approximately ten centuries ago, and it was extended to the occidental Mediterranean area by the Arabs [8]. Moroccan sorghum is essentially represented by the durra race [9], which is related for its expansion toward Asia and Mediterranean to the history of Islamisation [2,10].
In Morocco, sorghum is considered a minor cereal and cultivated on 29,000 ha, corresponding to 0.3% of the Moroccan arable surface [9]. Nowadays, this species is a culture regularly practiced in the northwestern regions of Morocco at altitudes of 20 to 800 m. In these regions, sorghum constitutes one of the most important cereals by the extent of its surfaces in comparison to corn, and by its food-producing character [9]. The Moroccan production fluctuates around 6 q/ha and never exceeds 9 q/ha [9]. These low yields are assigned to the traditional agricultural techniques based on old methods, lands parcelled out, and to the use of non-improved seeds. These seeds were not exchanged between regions, and they generate the local value of the ‘ecotype’ [9]. Thus, the knowledge of the genetic structure of these ecotypes is important, since these are populations of weak dimensions and in progressive regression. The phenotypic selection exerted by farmers has engendered landraces or locally adapted varieties. This selection has begun with the introduction of sorghum in Morocco and carried on about 900 to 1000 generations. Landraces are characterized by morphological differences (panicle, grain, awn, colour ...) that have been confirmed by several studies. A high degree of morphological variation among landraces, which were assigned to the race durra, bicolor and their intermediates, was noted [9]. Morphological characters and allozymes could account for 63% and 20%, respectively, of the variability between fields [11]. Morphological and enzymatic markers have been typically used to study the genetic variation of sorghum cultivars (e.g., [10–15]). Molecular markers constitute preferential tools to assess the genetic diversity and the phylogenetic relationships in sorghum; several studies have used the following analytical methods: RFLP markers [16–18], mitochondrial DNA markers [19], RAPD markers [20], and microsatellites markers [21,22].
In this investigation, we analyzed DNA bulks using RAPD and ISSR markers, which have never been used on the Moroccan sorghum. This approach allows us to get more genetic information on the studied populations. Our objective was to evaluate the genetic variability of northwestern fields, and to estimate the level of the variation between the studied regions. The obtained data were compared to those obtained with other markers (morphological traits, allozymes, and SSR) in the same regions.
2 Materials and methods
2.1 Plant materials
Seed samples were directly collected within fields (in situ sampling varying from 6 to 17 individuals) in four regions of northwestern Morocco: Larache, Tangier, Chefchaouen, and Tetouan (Fig. 1). Table 1 shows the plant material used for this study (accessions Lb1 to Teb5) and its geographical origin by region.
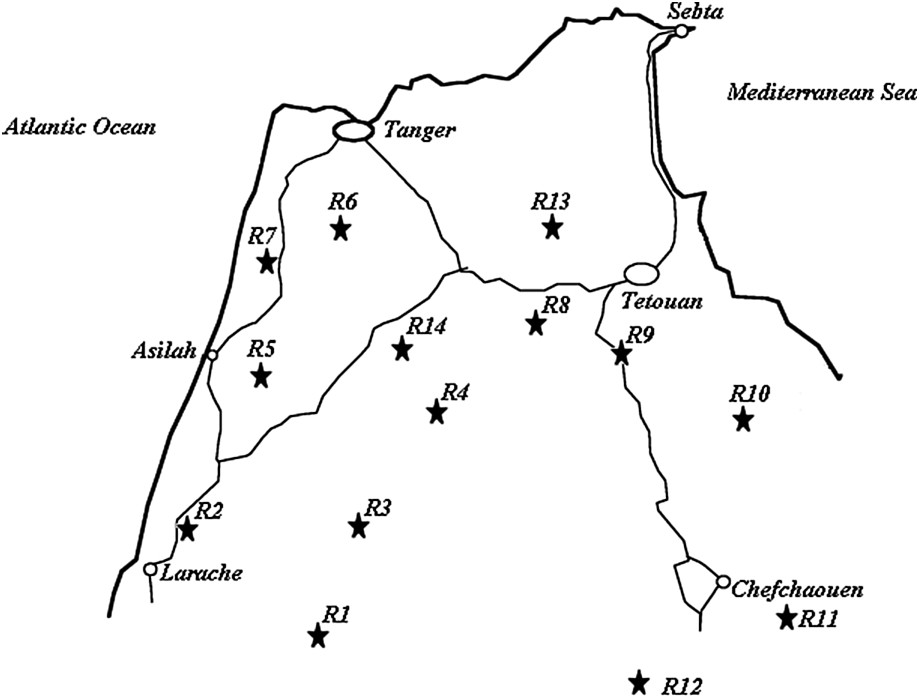
Geographical location of the studied sorghum cultivation regions in northwestern Morocco.
Accessions list and geographical origin of the Sorghum plant samples
| Accession (field) | Province | Region | Locality | Number of individuals per field |
| Lb1 | Larache | R1 | Oulad ben Lahcen | 12 |
| Lb2 | Naaimiine | 12 | ||
| Lb3 | Boujediane | 10 | ||
| Lb4 | Maâizate | 10 | ||
| Lb5 | R2 | Oulad Soltan | 12 | |
| Lb6 | Arrbaâ Ayacha | 10 | ||
| Lb7 | Douar hjrat el Garni | 10 | ||
| Lb8 | Douar Ain Mouloud | 10 | ||
| Lb9 | Mghazlienne | 10 | ||
| Lb10 | J'miaât 1 | 11 | ||
| Lb11 | J'miaât 2 | 11 | ||
| Lb12 | R3 | Luxus | 12 | |
| Lb13 | Khmis sahel/Arrihiyine | 13 | ||
| Tab1 | Tanger | R4 | Douar oulad larbi/Boutil | 13 |
| Tab2 | Takrii | 11 | ||
| Tab3 | M'nazla | 11 | ||
| Tab4 | R5 | Had Bghaghaz Dar chaoui | 10 | |
| Tab5 | R6 | Route Tanger-Tetouan1 | 10 | |
| Tab6 | Route Tanger-Tetouan2 | 10 | ||
| Tab7 | Zaînate | 15 | ||
| Tab8 | R7 | Route Asilah-Tanger | 4 | |
| Tab9 | Forêt diplomatique | 16 | ||
| Tab10 | Melloussa douar el Mekhfi | 10 | ||
| Tab11 | Douar Ain Homra | 12 | ||
| Chb1 | Chefchaouen | R8 | Ouad laou | 19 |
| Chb2 | R9 | Talembote | 6 | |
| Chb3 | R10 | Taghzoute | 15 | |
| Chb4 | R11 | Dardara1 | 16 | |
| Chb5 | Dardara2 | 17 | ||
| Teb2 | Tetouan | R12 | Beni Maaden | 11 |
| Teb3 | Douar Lhbar | 13 | ||
| Teb4 | R13 | Khemis Anjra | 17 | |
| Teb5 | R14 | Beni Harchin | 12 |
2.2 DNA extraction
DNA was extracted from a bulk of a totality of individuals for each field (population) as described by [23], with major modifications. Three grams of fresh leaves, stored at −80 °C, were ground in liquid nitrogen, mixed with 10 ml of CTAB extraction buffer [Tris 0.22 M pH 8.0, EDTA 0.22 M, Sorbitol 0.14 M, NaCl 0.8 M, N-laurylsarcosine 1% (w/v), CTAB 1.5% (w/v), and β-mercaptoethanol 1% (v/v), incubated at 55 °C for 1 h, and then cooled on ice and mixed with 5 ml of a phenol/chloroform/isoamylic alcohol (v/v, 25/24/1) mixture. The tube was centrifuged at for 5 min and the supernatant transferred into a new tube. The DNA was precipitated by cool isopropanol, washed with 75% ethanol, treated by the RNAse A and then by proteinase K before being reprocessed again to the phenol/chloroform/isoamylic alcohol mixture, and reprecipitated with NaCl and absolute ethanol. Centrifugation was performed at . The pellet was dried, resuspended in TE buffer [Tris-HCl 10 mM; EDTA 0.1 mM, pH 8.0]. DNA concentration and quality were determined through 0.8% agarose gel electrophoresis in 1X TBE buffer [Tris-HCl pH 8.0, 0.89 M; voric acid 0.89 M; EDTA 25 mM, pH 8.0] [24] with ethidium bromide staining, and by spectrophotometric measurements at 260 and 280 nm.
2.3 DNA amplification by RAPD and ISSR
The polymerase chain reaction (PCR) was run according to the modified protocol of [25] for RAPD and of [26] for ISSR, in a final volume of 25 μl containing 10 mM Tris-HCl pH 8.3 (at 25 °C), 50 mM KCl, 2.5 mM MgCl2, 2 mM aliquots of dATP, dCTP, dGTP, and dTTP (Promega), 0.2 μM RAPD primer or 4 μM ISSR primer, 100 ng of genomic DNA, and 1 unit of Ampli Taq DNA polymerase (Roche). The amplifications were performed on a GenAmp thermal cycler (Applied Biosystems, CA, USA) for RAPD following the program: at 95 °C for 5 min, 45 cycles (1 min at 95 °C, at 35 °C for 1 min, at 72 °C for 2 min) and final elongation of 6 min at 72 °C, and for ISSR: at 94 °C for 7 min, 32 cycles of 30 s at 94 °C, 45 s at 45 °C, 2 min at 72 °C, and final extension at 72 °C for 7 min. A total of 81 (10 bp) random primers (AD02, AK01-04, AM01-05, AN01-04, B10, C01-07, D01-05, E01-05, F01-05, G01-05, H01-05, I01-05, AF01-05, M01, R01-03, S01-08, T01-08, U02-03, U09 et Y15) (Operon Technologies, Almeda, CA, USA) were used in RAPD, and 4 primers F01: (CA)6AT, F02: (CA)6GC, F03: (CA)6AG and F04: (AGC)4T from (Operon Technologies) were used in ISSR. RAPD and ISSR amplification products were analysed by electrophoresis onto 1.2% and 2.8% agarose gels, respectively, run in 1XTBE and visualized by staining with ethidium bromide and illumination with UV light.
2.4 Data analysis
The parameter ‘polymorphism information content’ (PIC) was calculated according to reference [27]: , where n is the sample size, and is the frequency of the ith pattern revealed by the RAPD or ISSR primer j summed across all patterns revealed by the primer j.
The electrophoretic gel images were processed with the aid of the Gel Compar II software (version 2.5; Applied Maths, Kortrijk, Belgium). Using the same program, RAPD and ISSR bands were scored as 0 for absence and 1 for presence in each field. Jaccard distances [28], based on similarity index, were calculated, dendrograms were generated using Ward algorithm [29] and multidimensional scaling (MDS) was configured to facilitate the graphical visualization of the relationship between the formed groups [30].
3 Results
3.1 RAPD analysis
The number of polymorphic fragments per profile varied per primer from 11 (OPU2) to 27 (OPI2), and ranged from 138 to 3992 bp (Table 2). The minimum size difference between any of the two polymorphic products generated by a primer was approximately 20 bp. Overall, 812 (85%) polymorphic fragments were generated, and 140 (15%) fragments were common to all accessions studied. Among polymorphic fragments, 25 unique markers were detected (3%): six fragments were specific for Tangier fields (AF-2, 1559 bp; E-3, 804 bp; H-3, 966 bp; T-1, 1742 bp; T-6, 762 bp and T-6, 656 bp), three for Larache fields (AM-2, 1054 bp; T-1, 489 bp and T-6, 1566 bp), four for Tetouan fields (C-6, 823 bp; R-1, 761 bp; S-7, 234 bp and S-7, 138 bp) and twelve for Chefchaouen fields (D-3, 381 bp; H-5, 2208 bp; I-2, 2459 bp; I-2, 2008 bp; R-1, 1905 bp; R-2, 2321 bp; R-2, 1998 bp; S- 6, 2795 bp; S-7, 1540 bp; T-4, 2829 bp; T-4, 2004 bp and T-6, 1672 bp). The diversity (D) values varied from 0.86 to 0.97, with an average of (Table 2). In addition, high heterogeneity among fields was observed (Fig. 2). The genetic distances calculated from these data, and represented in a dendrogram, allow the probable relationships among the fields to be evaluated (Fig. 3). Six principal clusters were obtained. The first cluster was composed by two sub-groups: the first regrouped three fields of Larache and the second contained two fields of Tetouan and one field of Larache. The second cluster comprised two sub-groups: the first contained two fields of Larache, one field of Chefchaouen, and one of Tetouan. The second sub-group was represented by two fields of Chefchaouen. The third cluster included four fields of Larache. The fourth cluster was represented by three fields of Tangier, one of Larache, and one of Tetouan. The fifth one was composed by two sub-groups: the first one regrouped two fields of Larache and two fields of Chefchaouen, whereas the second sub-group comprised three fields of Tangier. Five fields of Tangier were found in the sixth cluster.
RAPD and ISSR markers information and diversity in 33 sorghum fields
| Primer | Molecular weight range, bp | Total number of markers | Number of markers | |||
| M | P | D | ||||
| RAPD | OPAF2 | 476–3535 | 20 | 3 | 17 | 0.9200 |
| OPAF3 | 300–2455 | 22 | 3 | 19 | 0.8674 | |
| OPAF4 | 395–3269 | 18 | 2 | 16 | 0.9438 | |
| OPAK4 | 234–2420 | 25 | 4 | 21 | 0.9627 | |
| OPAM1 | 341–2010 | 20 | 3 | 17 | 0.8785 | |
| OPAM2 | 212–1624 | 18 | 2 | 16 | 0.8741 | |
| OPAM4 | 304–2451 | 27 | 5 | 22 | 0.8996 | |
| OPAN3 | 355–2939 | 29 | 4 | 25 | 0.9730 | |
| OPB10 | 491–1851 | 16 | 3 | 13 | 0.8985 | |
| OPC1 | 194–1733 | 25 | 4 | 21 | 0.9526 | |
| OPC3 | 185–2232 | 31 | 5 | 26 | 0.9790 | |
| OPC6 | 541–3369 | 24 | 3 | 21 | 0.9021 | |
| OPD1 | 361–2304 | 30 | 4 | 26 | 0.9731 | |
| OPD3 | 381–2361 | 23 | 3 | 20 | 0.9524 | |
| OPD5 | 327–2523 | 28 | 5 | 23 | 0.9635 | |
| OPE1 | 361–3255 | 21 | 2 | 19 | 0.9569 | |
| OPE3 | 309–2323 | 27 | 4 | 23 | 0.9577 | |
| OPE4 | 494–2014 | 18 | 3 | 15 | 0.9276 | |
| OPF1 | 185–3104 | 25 | 4 | 21 | 0.9402 | |
| OPF3 | 419–3174 | 25 | 5 | 20 | 0.9525 | |
| OPF4 | 212–2951 | 24 | 2 | 22 | 0.9671 | |
| OPF5 | 333–3520 | 24 | 4 | 20 | 0.9329 | |
| OPG2 | 182–1920 | 26 | 3 | 23 | 0.9449 | |
| OPG3 | 447–3992 | 29 | 4 | 25 | 0.9618 | |
| OPH3 | 411–2100 | 26 | 3 | 23 | 0.9714 | |
| OPH5 | 241–2208 | 29 | 4 | 25 | 0.9767 | |
| OPI2 | 267–2752 | 33 | 6 | 27 | 0.9768 | |
| OPI3 | 189–2421 | 30 | 4 | 26 | 0.9629 | |
| OPR1 | 330–3100 | 28 | 5 | 23 | 0.9658 | |
| OPR2 | 358–2600 | 28 | 4 | 24 | 0.9633 | |
| OPS6 | 342–2795 | 23 | 3 | 20 | 0.9451 | |
| OPS7 | 138–1847 | 29 | 4 | 25 | 0.9717 | |
| OPT1 | 199–2380 | 28 | 3 | 25 | 0.9726 | |
| OPT4 | 195–2829 | 31 | 5 | 26 | 0.9744 | |
| OPT5 | 300–3293 | 28 | 3 | 25 | 0.9708 | |
| OPT6 | 415–2638 | 24 | 3 | 21 | 0.9518 | |
| OPU2 | 583–1940 | 15 | 4 | 11 | 0.9190 | |
| OPU3 | 233–1639 | 25 | 5 | 20 | 0.9539 | |
| Total | 952 | 140 | 812 | |||
| Polymorphism percentage | 85% | – | ||||
| ISSR | (CA)6AT | 314–2562 | 36 | 1 | 35 | 0.9855 |
| (CA)6GC | 183–1931 | 44 | 0 | 44 | 0.9989 | |
| (CA)6AG | 122–2317 | 50 | 1 | 49 | 0.9984 | |
| (AGC)4T | 160–1638 | 47 | 2 | 45 | 0.9964 | |
| Total | 177 | 4 | 173 | |||
| Polymorphism percentage | 98% | – |
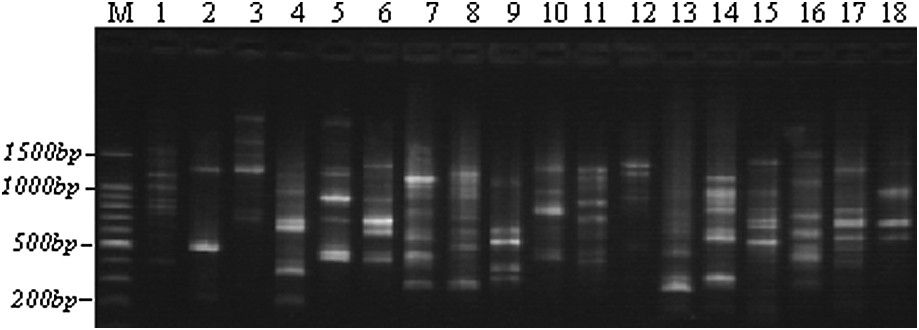
Example of an RAPD polymorphism obtained with the OPAN3 primer through TBE-agarose gel electrophoresis. M: 100-bp ladders; 1–5: Lb1, Lb3, Lb6, Lb8, and Lb10; 6–10: Tab1, Tab4, Tab6, Tab9, and Tab11; 11–14: Chb1, Chb2, Chb3, and Chb5; 15–18: Teb2, Teb3, Teb4, and Teb5.

Dendrogram obtained with Jaccard's similarity coefficients using RAPD data of 33 fields of Sorghum bicolor L. Moench (Lb1–13; Tab1–11; Chb1–5; Teb2–5). The tree was constructed using the Ward algorithm.
The spatial configuration of multidimensional scaling elaborated based on RAPD data allowed the distribution of the fields on three sectors according to the X-, Y-, and Z-axes to be established.
The (A) sector contained eight fields of Tangier and two fields of Larache, the (B) sector grouped six fields of Larache, three fields of Tangier, and two fields of Tetouan, and the (C) sector was represented by the entirety of Chefchaouen fields, five fields of Larache, and two fields of Tetouan (Fig. 4).
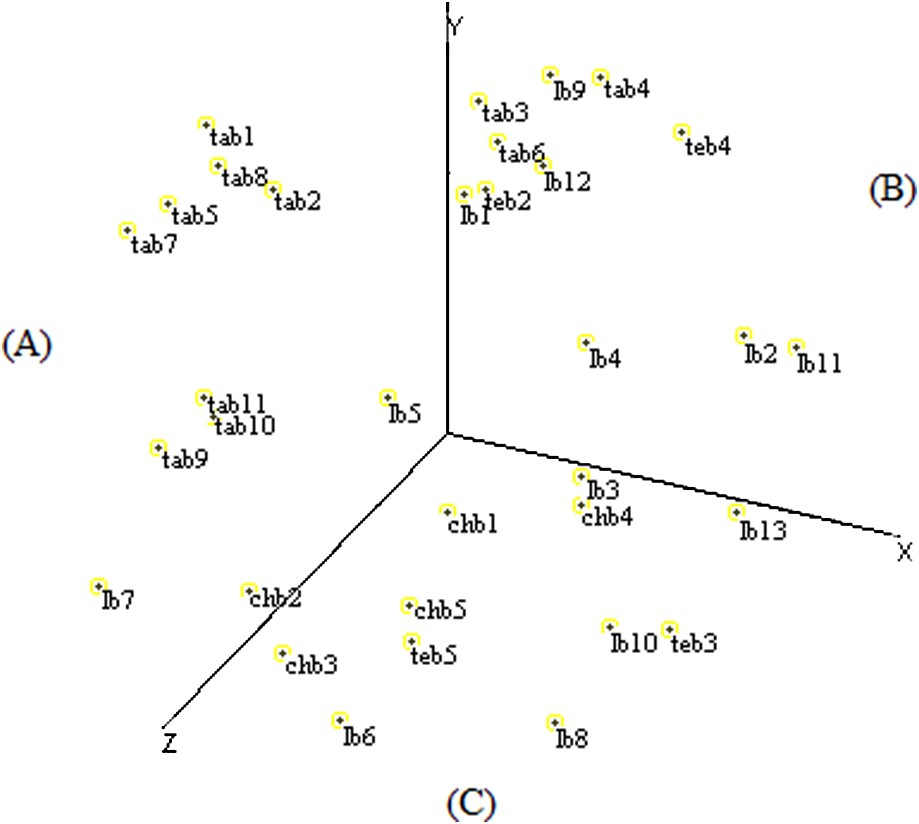
Scatter diagram of the MDS analysis of Moroccan sorghum accessions based on RAPD data.
3.2 ISSR analysis
The number of polymorphic fragments was important for the four primers, with an average of 43 bands, and ranged in size from 122 to 2562 bp (Table 2). The minimum size difference between any two polymorphic products generated by a primer was approximately 10 bp. Generally, 173 (98%) polymorphic fragments were generated, and 6 (2%) fragments were common to all accessions. Among polymorphic fragments, four different classes of markers were distinguished: (1) markers that were found in the four studied regions – Tangier, Larache, Chefchaouen and Tetouan (39%) –; (2) markers that were common only between the fields of Larache and some fields of Tangier (33%); (3) markers that were common between both fields of Tetouan and Chefchaouen and some fields of Tangier (10%); (4) a class of 31 unique markers (18%). Fourteen fragments were specific to the Larache fields (F-1, 715 bp; 534 bp; 320 bp; F-2, 715 bp; 534 bp; 183 bp; F-3, 1459 bp; 675 bp; 568 bp; 217 bp; 174 bp; 167 bp; F-4, 1638 bp; 161 bp), 13 to the Tangier fields (F-1, 2562 bp; 1170 bp; 919 bp; 359 bp; F-2, 1458 bp; 1369 bp; 989 bp; 376 bp; 330 bp; F-3, 2317 bp; 209 bp; F-4, 1638 bp; 161 bp), two to the Chefchaouen fields (F-1, 631 bp and F-4, 209 bp), and two to the Tetouan fields (F-3, 141 bp and F-4, 466 bp). The diversity (D) values varied from 0.98 and 0.99, with an average of . High heterogeneity among fields was observed and each field was distinguished (Fig. 5). The genetic distances were calculated from the obtained data, allowing the probable relationships among the fields to be evaluated (Fig. 6). Two principal clusters were obtained. The first cluster was composed of two sub-groups: the first one regrouped eight fields of Larache and two fields of Tangier, whereas the second sub-group was divided into two parts, the first regrouping four fields of Tangier and the second five fields of Larache associated with one field of Tangier. The second cluster was composed by two sub-groups; the first one was divided into three parts: the first part included three fields of Chefchaouen, the second part contained three fields of Tetouan, and the third part regrouped two fields of Chefchaouen and one field of Tetouan; the second sub-group was represented by four fields of Tangier.
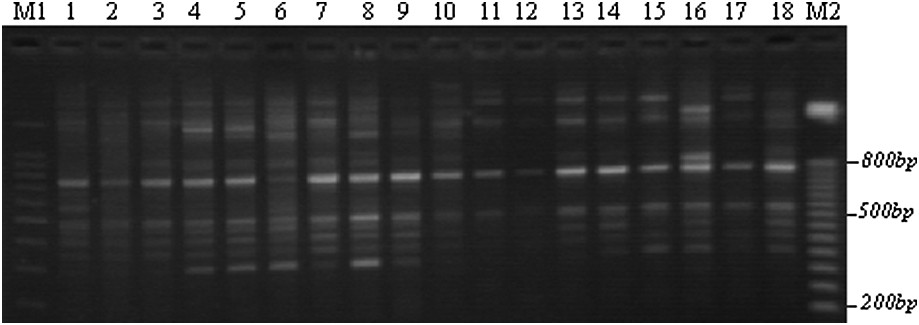
Example of an ISSR polymorphism obtained with the F1 primer through TBE-agarose gel electrophoresis. M: 100-bp DNA ladders; 1–5: Lb1, Lb3, Lb6, Lb8, and Lb10; 6–10: Tab1, Tab4, Tab6, Tab9, and Tab11; 11–14: Chb1, Chb2, Chb3, and Chb5; 15–18: Teb2, Teb3, Teb4, and Teb5; M2: 50-bp DNA ladders.

Dendrogram obtained with Jaccard's similarity coefficients using ISSR data of 33 fields of Sorghum bicolor L. Moench (Lb1–13; Tab1–11; Chb1–5; Teb2–5). The tree was constructed using the Ward algorithm.
The distribution of fields by MDS based on ISSR data determined three sectors according to the X-, Y-, and Z-axes. The (A′) sector contained nine fields of Larache and five fields of Tangier, the (B′) sector grouped five fields of Tetouan, two fields of Tangier and one field of Chefchaouen, and the (C′) sector was represented by four fields of Chefchaouen, four fields of Larache, and four fields of Tangier (Fig. 7).
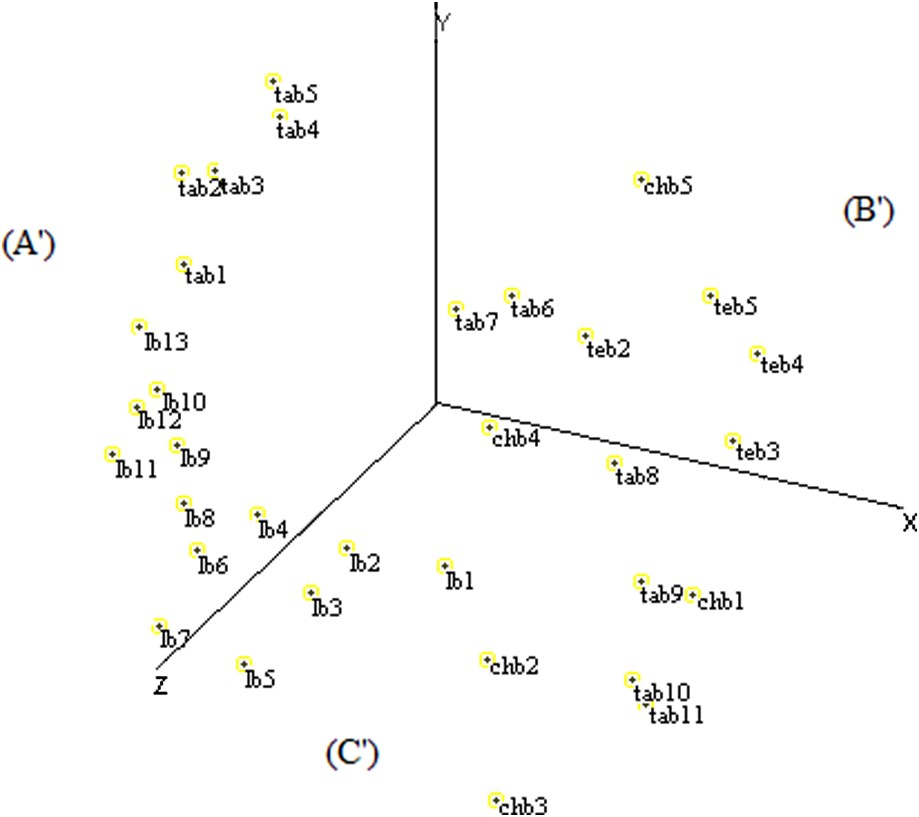
Scatter diagram of the MDS analysis of Moroccan sorghum accessions based on ISSR data.
4 Discussion
DNA analysis using RAPD and ISSR markers on seed bulks was a useful tool to assess the genetic diversity of the Moroccan populations of Sorghum bicolor L. Moench. A great number of bands were obtained with each primer and for the two types of markers. The totality of RAPD and ISSR primers were highly informative, PIC values were close to 1 [27]. The variability level between the different fields revealed by both RAPD and ISSR is very important. Our results are in keeping with the works of Djè et al. [11], who showed a large proportion of morphological variability between fields, while allozymes and microsatellites revealed more intra-field than inter-field variability [21].
Additional region-specific polymorphic fragments were found in this study using the RAPD and ISSR techniques. RAPD revealed 13 specific markers for Larache, 6 for Tangier, 4 for Tetouan and 12 for Chefchaouen. ISSR revealed 14 specific markers for Larache, 13 for Tangier, 2 for Tetouan, and 2 for Chefchaouen. These results suppose the probable presence of specific genomic zones that are common to the totality of the studied populations belonging to the same region. These methods revealed more diversity than the use of morphological traits, allozymes, or microsatellites previously used to distinguish between regions [9,19]. The specificity of the ISSR allowed us to divide studied fields into two groups according to two classes of markers: the first ones are common to the fields belonging to the west of Tangier and all fields of Larache, and the second ones are common to the fields belonging to the east of Tangier and to all fields of Tetouan and Chefchaouen. These data show that there exists an influence of the micro-geographical distribution of sorghum fields on the structuring of their variability, while Djè et al. [11] have also demonstrated that there was no correlation between geographical distances and micro-geographical and allozymic data. Our results confirm those found by Djè et al. [12], who showed that quantitative characters, in contrast to qualitative traits, are influenced by location, topography, and soil fertility.
Compared to RAPD, the clustering and multidimensional scaling obtained by ISSR groups more clearly defines fields according to their geographic distribution. The flexible reproduction system of the Moroccan sorghum combining autogamy and allogamy supposes genes flow between fields to be in favour of a similarity between nearby fields and therefore between fields belonging to the same region [31]. Our results are confirmed by data obtained with the morphological analysis carried out by Kadiri and Ater [9], who found that Moroccan sorghum would belong to two cultivars of the race durra, durra, and cernuum, which occupied two domains of distribution that are geographically different. The first domain is internal and the second domain is coastal external of the Rifian chain. Moreover, the first field of Larache (lb1, Ouled ben Lahcen) was isolated in both ISSR and RAPD clustering. This can be explained by the introduction of seeds of American cultivars during trials [9].
The local seeds of the sorghum constitute veritable populations qualified as ecotypes that have resulted from a long process of selection carried out by the farmer in order to choose adapted plants. The polymorphism was important; therefore, each population represents a conservation entity and constitutes an important reserve of genes to be used for the improvement of this cereal. These ecotypes have very interesting potentialities for their hardiness and their capacity to give constant yields despite the environmental conditions. Thus, all efforts to evaluate this potential would be important and urgent.
Acknowledgements
This work was supported by the Moroccan Ministry of the National Education, Scientific and Technical Research, PROTARS II-P52/03 (2002–2006), and was also helped by the Organization of the Islamic Conference (OIC). We are especially grateful to Sir A. Belkeziz, and thankful to Dr. A. Sasson for his encouragements, assistance, and counsels. We thank Laila Sbabou for her assistance and for reviewing the manuscript.


