1. Introduction
Macromolecular storage is an interesting emerging trend for saving data at the molecular scale [1, 2, 3]. In this approach, molecular building blocks (i.e. monomers) are set as basic information units and their arrangement in a polymer chain is exploited as a processable (i.e. writable, readable and editable) information string. This can be achieved with biological informational polymers, such as DNA [4, 5], but also using a wide variety of synthetic copolymers, as demonstrated by our group [6, 7, 8, 9] and others [10, 11, 12, 13]. Since academic research about automated chemical synthesis (i.e. writing) [14] and automated sequencing (i.e. reading) [15] of DNA is much more mature than the one about synthetic informational polymers, DNA data storage currently gives access to much higher storage capacities than synthetic analogues [5]. Yet, it has been recently proposed that the storage properties (e.g. density, capacity and stability) of synthetic polymers might overrule those of DNA on the long term [16]. Indeed, the molecular structure of synthetic macromolecules can be varied almost infinitely to attain optimal properties, whereas the one of DNA is fixed by biological constraints.
Among the different types of synthetic polymers that have been reported for data storage, abiotic poly(phosphodiester)s are a very promising option [16]. These sequence-defined macromolecules are synthesized by automated phosphoramidite chemistry; an approach that is also used for the chemical synthesis of DNA [17]. However, non-natural monomers are used in these syntheses instead of nucleoside phosphoramidites [18]. Consequently, in terms of molecular structure, these poly(phosphodiester)s have nothing in common with nucleic acids at the exception of phosphates formed by phosphitylation and subsequent oxidation. Hence, these polymers allow information storage but their molecular structure is not restricted by biological constraints. Over past years, we have therefore gradually improved their design to render them more and more suitable for data storage applications. The first generation of digital poly(phosphodiester)s was prepared in 2015 using a binary alphabet (i.e. two different monomers allowing a maximum storage density of 1 bit/monomer) [19]. Initially, the polymers were synthesized manually but later that year, we reported the automated synthesis of longer chains [20]. However, this first generation of digitally-encoded poly(phosphodiester)s was very difficult to decrypt using sequencing tools such as tandem mass spectrometry (MS/MS) [21] or nanopore sequencing [22, 23]. This issue was solved in 2017, when we reported the design of long poly(phosphodiester)s that can be decoded by MS/MS using a routine mass spectrometer [24]. To do so, tetramethylpiperidinyloxy (TEMPO)-based alkoxyamine motifs were periodically included in the polymer chains using an appropriate phosphoramidite building block. When subjected to MS/MS conditions, these sites break preferentially, thus leading to a library of coded fragments of defined size. Each fragment is pre-labelled with a mass-tag that permits its identification and therefore, after performing the sequencing of all fragments in pseudo-MS3 conditions, the complete information sequence can be recovered. Yet, a poly(phosphodiester) with a maximum storage capacity of 77 bits/chain was achieved in this work and its decryption required a time-consuming manual interpretation of the MS/MS and MS3 spectra. Although a software named MS-DECODER was developed the same year for the decoding of synthetic digital polymers [25], it could not be applied to these optimized poly(phosphodiester)s because TEMPO-based alkoxyamines lead to intense side peaks that hinder automated decryption. This problem was solved in 2020 using an optimized alkoxyamine motif named RISC2 (RISC stands for Ring InSide Chain) that minimizes side-products formation [26]. In parallel, we also reported in 2020 expanded monomer alphabets containing 4- or 8-symbols, which allow storage densities of 2 or 3 bits/monomer, respectively [27]. All these recent developments enable the design of high-capacity digital polymers. However, at present, the biggest digital poly(phosphodiester) ever decrypted had a storage capacity of 144 bits/chain, which is still far from the maximum possible. For instance, no compression algorithm was used to encode this polymer and its decoding was not performed automatically.
In this context, we report herein the synthesis and automated decryption of a poly(phosphodiester) of very high storage capacity. This was achieved by combining the four following solutions: (i) using an expanded monomer alphabet of 8-symbols, (ii) using the optimized alkoxyamine RISC2, (iii) developing an appropriate and expanded set of fragment markers, and (iv) using a compression algorithm. In order to illustrate the efficacy of this design, we describe here the preparation of a digital macromolecule that stores the portrait of the most renowned French Chemist, Antoine de Lavoisier (also known as Antoine Lavoisier after the French revolution). A pixelated version of a known engraving of Lavoisier was created, compressed and stored in a digital poly(phosphodiester). The single chain capacity of 440 bits/chain attained in this work is the highest ever reported for a synthetic informational polymer.
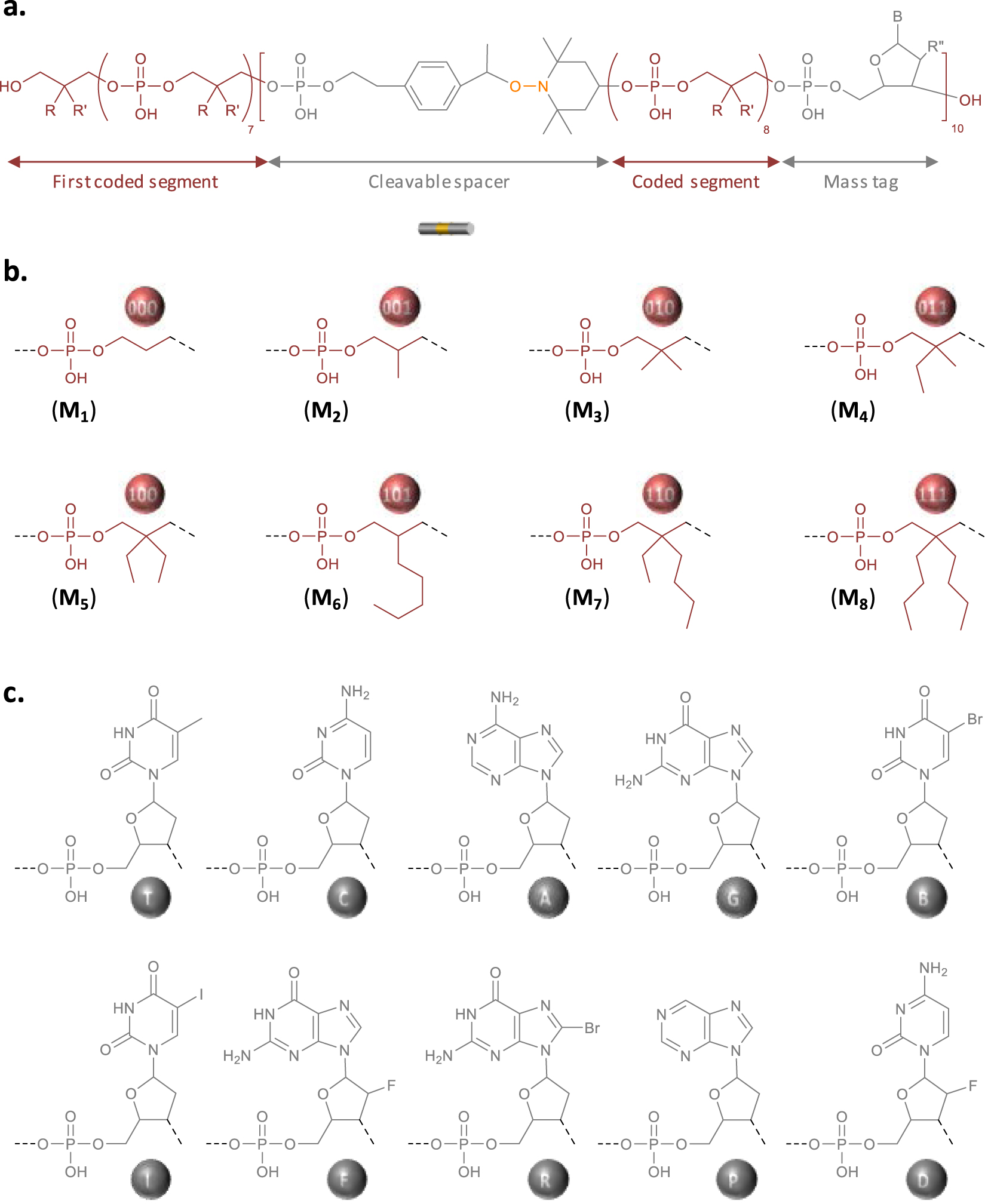
Design of the digital polymer. (a) General molecular structure of the digitally-encoded poly(phosphodiester) synthesized in this work. (b) Molecular structure of the eight different synthons that permit to code binary information in the chains. (c) Molecular structure of the ten different mass tags, which facilitate the decryption of the digital sequence by mass spectrometry.
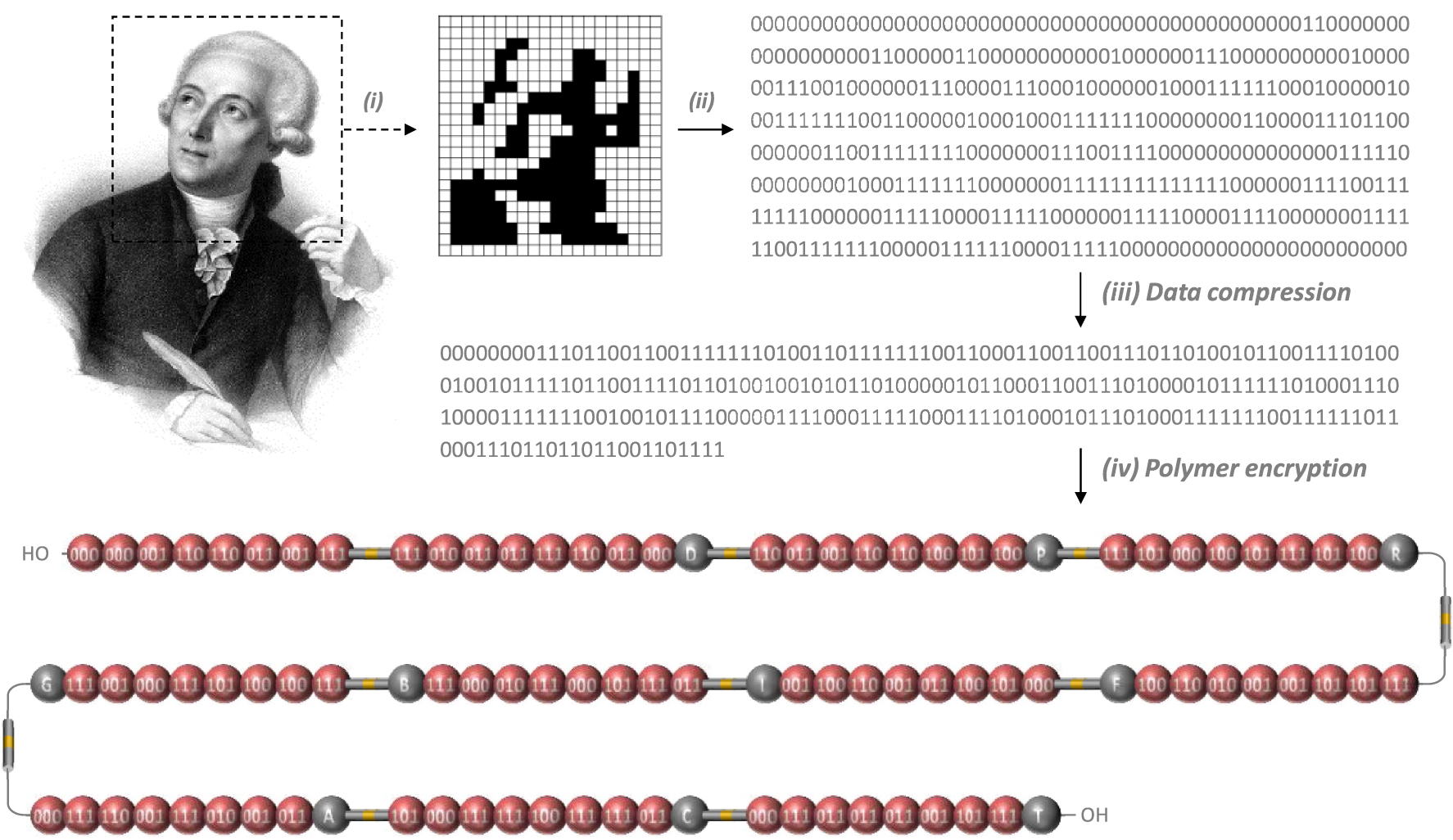
Polymer encryption. (i) The portrait of Lavoisier (Author: François Séraphin Delpech, public domain) was first pixelated into a 20 × 22 image (440 pixels). (ii) The pixels were then transformed into a 440-bits string with 0 and 1 coding for white and black pixels, respectively. (iii) This digital sequence was then compressed into a 264-bits string. (iv) The compressed sequence was then translated into a chemical monomer sequence employing the building blocks shown in Figure 1.
2. Results and discussion
The polymer studied in this work was synthesized by solid-phase phosphoramidite chemistry on an automated DNA synthesizer [14]. Synthesis was performed on a crosslinked polystyrene resin. In order to prepare a high-capacity digital polyphosphodiester, three types of phosphoramidite building blocks are necessary: (i) coded comonomers that allow information storage [19], (ii) an alkoxyamine-containing linker that guides fragmentation in MS/MS sequencing [24], and (iii) mass tags that permit to identify mass spectrometry fragments [24]. Figure S1 shows the molecular structure of all the phosphoramidite building blocks used in this work and Figure 1 displays the general molecular structure of the resulting poly(phosphodiester). Digital-encoding was achieved using an alphabet composed of eight different monomers (M1–M8 in Figure S1 and Figure 1) [27]. For instance, M1, M2, M3, M4, M5, M6, M7, and M8 code for the triads 000, 001, 010, 011, 100, 101, 110 and 111, respectively. As mentioned in the introduction, the recently-reported RISC2 compound was used in this work as alkoxyamine-containing linker [26]. It was included periodically in the chain (i.e. every eight coded monomers) as shown in Figure 1. Furthermore, ten different nucleosides were used as mass tags (T, C, A, G, B, I, F, R, P, D in Figure 1). Seven of them have already been used in previous works [24], while the other three, namely R, P and D, have been investigated for the first time in the present study. In order to enable the unambiguous identification of a chain-fragment, each mass tag shall have an exact molar mass that satisfies strict criteria, as previously described [24]. The new markers R, P and D were therefore carefully selected according to these rules. All the mass tags were incorporated in the polymer chains using the corresponding phosphoramidite monomers (Figure S1) with the exception of T that comes from the preloaded polystyrene support.
This molecular alphabet was used herein to synthesize a macromolecule that stores the pixelated portrait of Antoine de Lavoisier (Figure 2). The black & white Lavoisier picture was coded on a 20 × 22 grid (440 bits) with 1 bit coding for black pixels and 0 for white pixels. The picture, considered as a bit stream, was coded using an arithmetic coding compressing scheme developed by “project Nayuki” (code available in supporting information document) [28]. The series of 440 bits was first linearized and a checksum bit was added to it, and zero-padded to a length multiple of 8. Then a frequency table for 8 bits sequences was computed and used for arithmetic coding to compress the bit stream to 264 bits. This compressed sequence was then expressed as a sequence of 88 coded monomers, 10 alkoxyamine spacers and 10 mass tags (108 building blocks in total), as shown in Figure 2. Following previously established-conventions [20, 24, 27], the reading direction of the polymer was set opposite to the synthesis direction. The mass tags sequence is therefore read in the following order (from left to right in Figure 2): no tag, D, P, R, F, I, B, G, A, C, T. Following the acronym conventions set in Figure 1, the primary structure of the synthesized polymer is as follow: M1⋅M1⋅M2⋅M7⋅M7⋅M4⋅M2⋅M8-RISC2-M8⋅M3⋅M4⋅M4⋅ M8⋅M7⋅M4⋅M1⋅D-RISC2-M7⋅M4⋅M2⋅M7⋅M7⋅M5⋅M6⋅ M5⋅P-RISC2-M8⋅M6⋅M1⋅M5⋅M6⋅M8⋅M6⋅M5⋅R-RISC2- M8⋅M6⋅M6⋅M2⋅M2⋅M3⋅M7⋅M5⋅F-RISC2-M1⋅M6⋅M5⋅ M4⋅M2⋅M7⋅M5⋅M2⋅I-RISC2-M4⋅M8⋅M6⋅M1⋅M8⋅M3⋅ M1⋅M8⋅B-RISC2-M8⋅M5⋅M5⋅M6⋅M8⋅M1⋅M2⋅M8⋅G-RISC2-M1⋅M8⋅M7⋅M2⋅M8⋅M3⋅M2⋅M4⋅A-RISC2-M6⋅ M1⋅M8⋅M8⋅M5⋅M8⋅M8⋅M4⋅C-RISC2-M1⋅M8⋅M4⋅M4⋅ M4⋅M2⋅M6⋅M8-T.
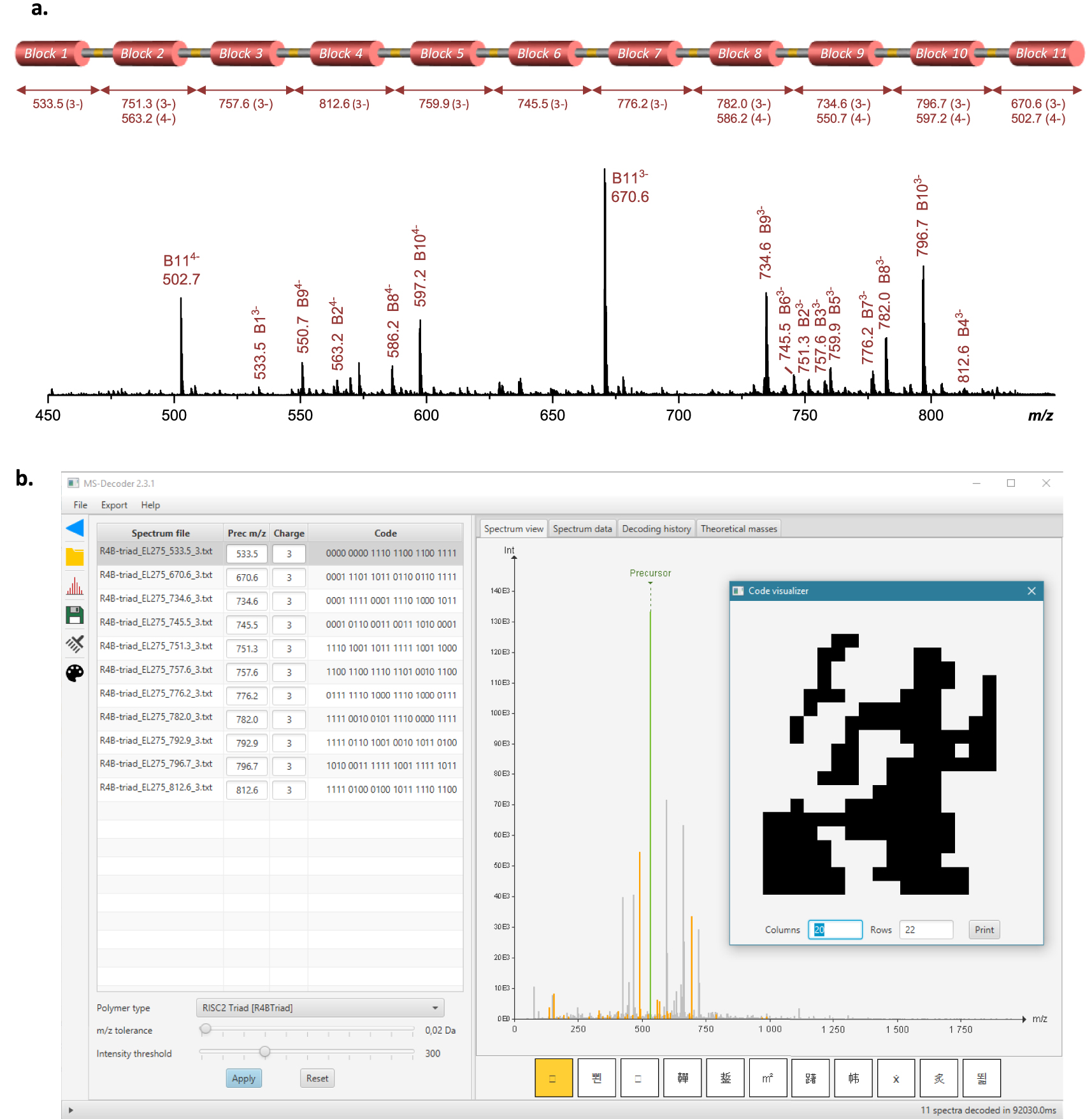
Polymer decryption. (a) ESI-MS spectrum recorded for the Lavoisier-containing polymer in negative mode, using a cone voltage of 60 V to induce cleavage of all alkoxyamine bonds. (b) Screenshot of the MS-DECODER interface for sequence annotation and reading.
The formed Lavoisier-containing polymer was characterized by size exclusion chromatography (SEC), electrospray ionization mass spectrometry (ESI-MS) and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS). The complete polymer (24 408 Da at isotopic maximum) was not detected by ESI-MS. Instead, a series of fragments resulting from a premature fragmentation of some alkoxyamine sites was observed, even when using the lowest cone voltage for smooth ion transfer from the atmospheric pressure source to the vacuum side of the mass spectrometer (data not shown). This behavior is most probably due to intense repulsion forces between numerous negative charges expected in the multi-deprotonated macromolecule (on average, 3 charges per coded segment) leading to in-source fragmentation when this macro-anion is accelerated through the medium pressure interface of the mass spectrometer. However, MALDI mass analysis allowed detection of the intact macromolecule as a singly deprotonated species (Figure S2). The high laser fluence requested to desorb the polymer from the MALDI sample also induced alkoxyamine bond cleavage, leading to successive release of coded segments hence providing additional evidence of the accurate structure (Figure S2). SEC analysis also showed a relatively well-defined macromolecule (Mn = 12,000 g⋅mol−1, = 1.25). Still, the refractometer signal exhibits low molecular weight shoulders that might be due to imperfect synthesis or partial polymer degradation (Figure S3). Nevertheless, the maximum peak value of the refractometer trace (Mp = 16,000 g⋅mol−1) indicates that the main population of the multimodal distribution has a molecular weight which is close to the expected theoretical value. Of note, the dn∕dc of the recorded polymer was only roughly estimated and therefore the measured molecular weight values are not meant to be absolute. Overall, ESI-MS, MALDI-TOF-MS and SEC results tend to indicate that the targeted polymer was synthesized, even though it is not pure. All signature fragments of the targeted sequence were confirmed by ESI-MS experiments performed with the cone voltage set to 60 V to induce cleavage of all alkoxyamine bonds: as shown in Figure 3a (and Table S1), the eleven coded blocks of the polymer were all individually observed in ESI-MS. This proves undoubtedly that the targeted primary structure was prepared. Consequently, all the sequence fragments were then subjected to further CID fragmentation and individually sequenced (Figures S4–S7)1. It shall be remarked that the polydispersity of the initial polymer does not affect sequencing because the ion sampled for fragmentation is selectively chosen based on its m∕z value. The obtained spectra were analyzed both manually and by the MS-DECODER software [25]. For this, the algorithm of the software was upgraded for decoding the 8-symbols alphabet and the 11 mass-tags that were used in this work. Overall, the complete information sequence was deciphered in about one minute, which is drastically faster than manual sequencing. During analysis, the bit stream to be decoded was determined from the observed monomer sequence, the frequency table was used to decompress the message, and the checksum bit was verified. The 440 bits thus obtain can then be displayed as a 20 × 22 picture (Figure 3b).
3. Conclusion
In summary, the synthesis and mass spectrometry decoding of an informational poly(phosphodiester) allowing an unprecedented storage capacity of 440 bits/chain was described in this work. This was attained by optimizing the molecular design of the polymer and by employing an appropriate compression algorithm. In terms of polymer design, a set of nineteen building blocks (eight coded monomers, ten mass tags and one cleavable spacer) was used to synthesize this polymer. The polymer was then decoded by multistage mass spectrometry. The intact macromolecule could be observed in MALDI-MS as a singly charged macro-anion but the highly charged species generated in ESI dissociated during its transfer in the vacuum side of the instrument. Nevertheless, the library of spontaneously-formed in-source fragments confirmed that the polymer was synthesized and allowed complete deciphering of the information sequence. Furthermore, thanks to the use of the optimized alkoxyamine spacer RISC2, the reading of the polymer could be automated using the MS-DECODER software. As a proof-of-principle, the portrait of Antoine de Lavoisier was stored at the molecular scale in the present work. Overall, this study is an homage to one of the founding fathers of modern chemistry and underlines that the limits of chemically-synthesized informational polymers have not yet been reached [30, 31].




 CC-BY 4.0
CC-BY 4.0