1 Introduction
Astringency is considered as a proof of red wine quality. It appears as a dry, rough or even pucker sensation in the entire mouth while tasting [1]. A consensus of opinion suggests that this sensation is the result of a recognition process occurring between tannins and saliva proteins [2–8]. Wine tannins are mainly issued from the solid part of the grape, skins, seeds and stems. They are transferred to wine during wine-making and can reach a concentration up to 4 g/l in Bordeaux red wines. They are oligomers of flavan-3-ol also called Procyanindins [9] and represent a large family owing to their two distinct constitutive units, the two types of linkage possible in between two units (4-6 or 4-8) and the number of constitutive units or degree of polymerization (up to 13 constitutive units were found in red wine [10]).
Human parotid saliva proteins are composed up to 70% of Proline-Rich Proteins (PRPs) divided in three groups depending on their specific functions, glycolysed, acid and basic PRPs [11]. Only basic PRPs are known to bind polyphenols [12,13] and this specific binding results in the tactile astringent sensation. As their name let us suggest, PRPs are mostly composed of Proline (40%) but also of glycine and glutamine, these three amino acids corresponding up to 80% of the total amino acids content [14]. They are organized in two types of repeated sequences of five (PQGPP) and nine amino acids (PPGKPQGPP), which confer on them an extended structure open, to interact with tannins [15]. In the present work, we monitor the interactions of the four different 4-8 linked procyanidin dimers (Scheme 1) with a peptide of 14 residus (IB7-14) representing a PRP widespread consensus sequence using different types of NMR experiments. Following the peptide proton chemical shift or even the diffusion coefficient variations induced by adding the different procyanidins gives rise to some physico-chemical parameters of interest [16]: the stoichiometry of the complex formed, the interaction sites and the affinity constant of the complex formation. The differences observed in between tannins could be used to establish an affinity scale of tannins, based on the hypothesis that the astringency sensation is directly linked to the affinity of a tannin towards a saliva PRP.
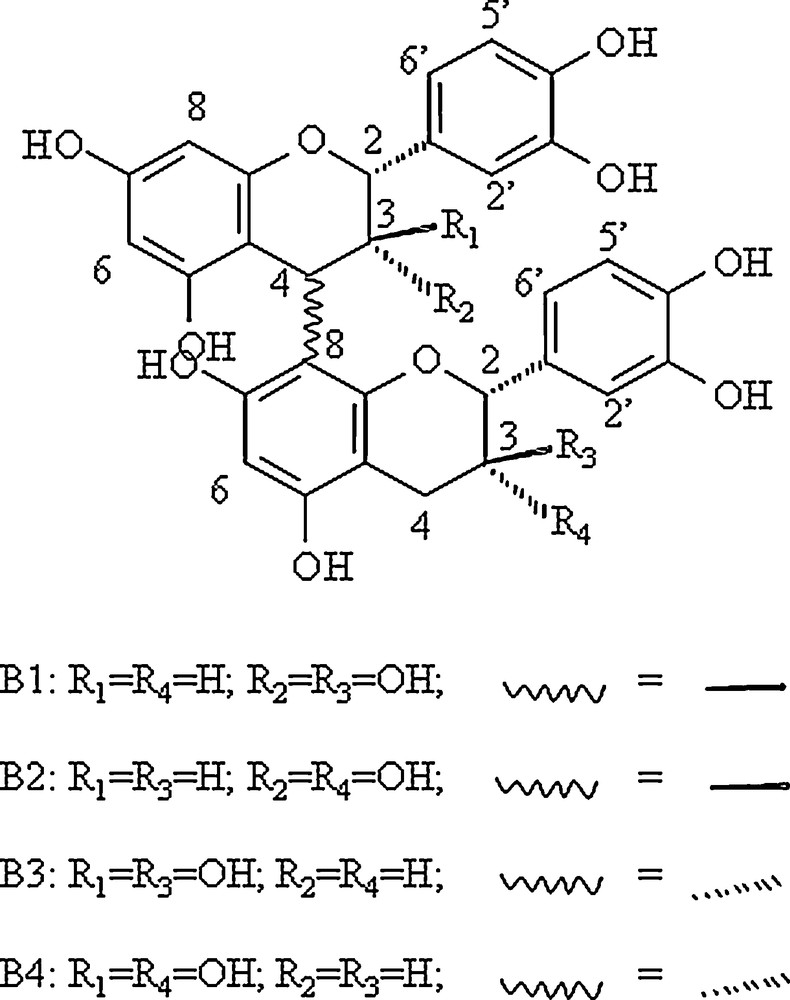
Structure of the 4 4-8 procyanidin dimers studied.
2 Materials and methods
2.1 Synthesis
The chemical synthesis of the peptide IB7-14 (SPPGKPQGPPPQGG) was performed on an Applied Biosystems petide synthetizer 433A by the FastMoc methods using a method previously described [17]. Procyanidins dimers B1, B2, B3, and B4 (Scheme 1) were synthesized using the previously described methods [18].
2.2 Sample preparation
A 1 mM protein sample was dissolved in a water/D2O/EtOD: 80/8/12 mixture, with 5 mM deutered acetic acid, pH 3.5 to mimic wine conditions. Tannins were progressively added to the protein sample to scan a tannin concentration ranging from 1 to 7 mM. In order to keep constant the protein concentration, tannins were just added under a powder form after being dissolved in water, aliquoted and lyophilized. Using such a procedure, the same 5 mm NMR tube was used with the 1 mM protein solution and lyophilized tannin was added for each ratio.
2.3 NMR experiments
Spectra were recorded at 298 K on a Bruker DXP 400 spectrometer equipped with a 5 mm gradient inverse broadband probe with 2H lock. All proton chemical shifts are given with respect to TSP as an internal reference so that it also can be used for quantification purpose. 1D spectra were recorded using a Watergate Sequence in order to suppress efficiently the water resonance [19]. Diffusion measurements were performed using the Stejkal and Tanner pulse sequence [20] using the following parameters: spectral width = 12 ppm; NS = 64; recycling delay = 3 s; intergradient delay Δ = 150 ms; gradient pulse duration δ = 2 ms. The pulse gradients (G) were incremented from 2 to 95% of the maximum gradient strength in a linear ramp in 16 steps. The diffusion coefficient, D, can be obtained by fitting a specific resonance area, I, obtained at different gradient powers (G) using the following equation:
I = e− γ2G2Dδ2 (Δ−δ/3).
NMR data analysis: For titration experiments, chemical shifts variations of some protons of the peptide were analyzed as a function of procyanidin dimer concentration using the equation previously described for a multisite model [21]:
| (2) |
where Aobs is Δδi, the chemical shift variation (ppm) or ΔDobs, the diffusion coefficient variation, Amax is Δδmax (or ΔDmax), the chemical shift (diffusion D) difference between the chemical shift (or D) of the protein alone and the protein saturated in tannins, Kd is the dissociation constant expressed in M, [Ti], the total concentration of polyphenol able to fix the peptide (M) by taking into account their autoassociation (the Ka of the autoassociation process has been evaluated at around 6 M−1, [22]), [P0], the total concentration of peptide (M), and n, the number of polyphenol binding sites. Kd, n and Δδmax were calculated using a least-squares-fitting routine within the software program EXCEL.
3 Results
3.1 Chemical shift variations
Fig. 1 shows the chemical shift variations of the peptide NH resonances when procyanidin B1 is progressively added to the peptide solution. All the peptide NMR proton signals have been already assigned [16]. Significant chemical shifts are observed for all the NH resonances but the effect is exemplified on the resonances of Glycine 13 and 14 (G13, G14) NH. Proline 2, 9 and 10 (P2, P9, P10) Hα resonances are also affected by the presence of procyanidin dimers. This suggests that the fixation sites are probably the same for each tannin. These chemical shift variations were fitted using Eq. (2) describing the fixation process of n tannins on a protein with the same affinity (i.e., the same Kd for every tannin), as previously described [23]. Fig. 2 displays G13 NH chemical shift variations observed when B1 (diamond), B2 (triangle), B3 (circle) or B4 (square) are progressively added (ranging from 0 to 7 mM) to the 1 mM peptide solution, and their fitting (lines), Kd, n, and Δδmax being the adjustable parameters. Kd and n values obtained for different protons (G13 and G14 NH, P2, P9 and P10 Hα) were averaged and are reported on Table 1.
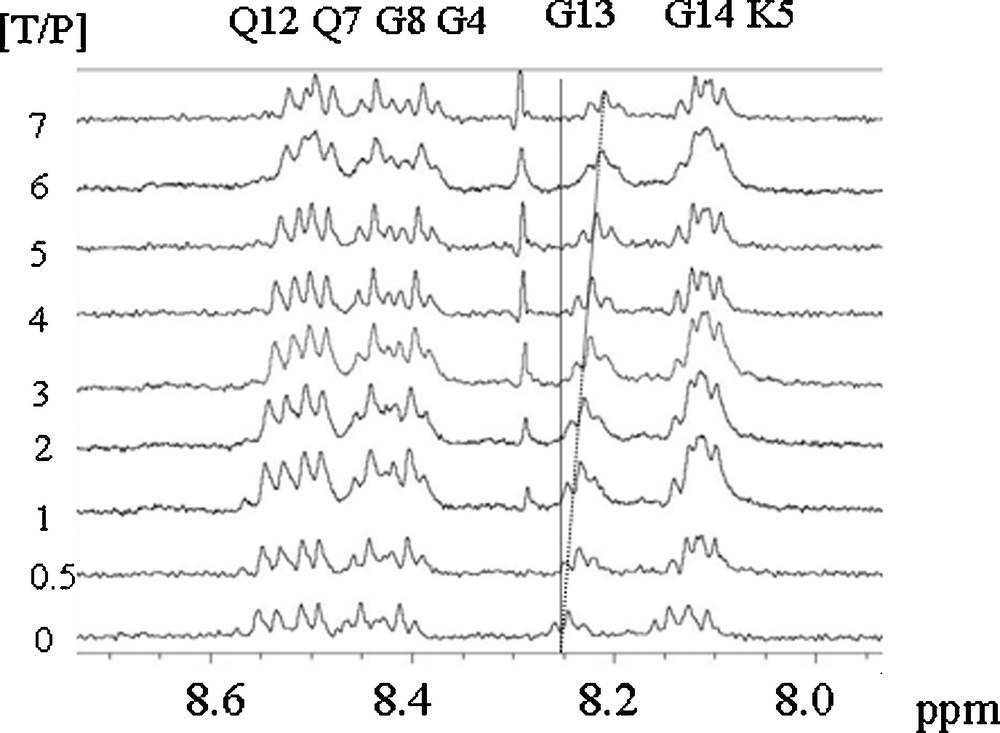
1H spectra of a 1 mM peptide IB7-14 solution recorded with different procyanidin contents (from 0 to 7 mM).
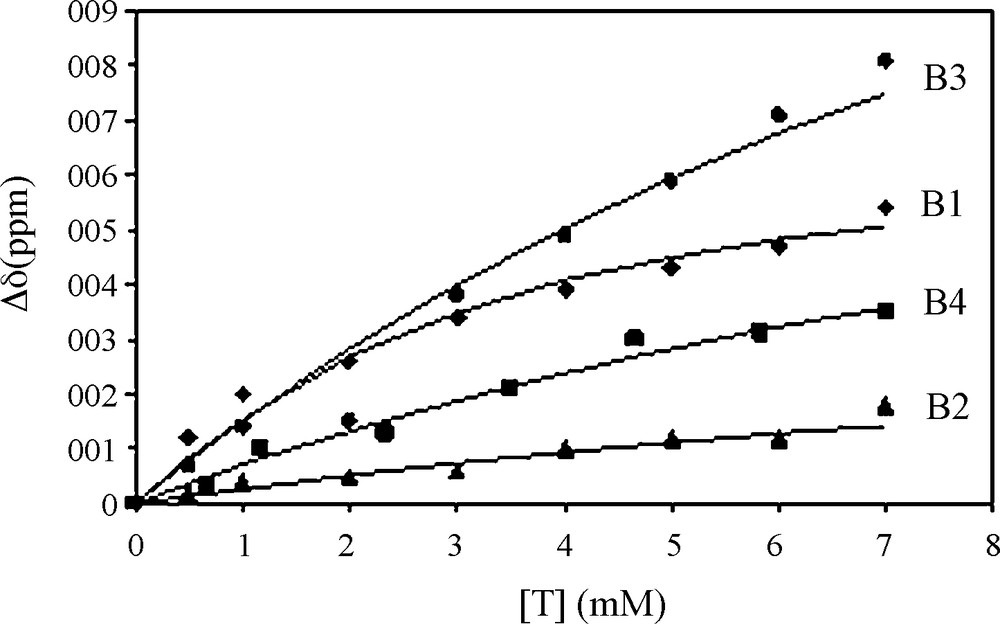
Chemical shift variation of G13 NH of the peptide (1 mM) when different procyanidins are added.
Dissociation constant (Kd), and number of tannin binding sites (n) were obtained from the fit of experimental chemical shift variations of G13 and G14 NH, P2, P9 and P10 Hα, and from the diffusion coefficients variation of the petide using eq. (2). The different Kd and n values obtained for one tannin were averaged are reported ± SD.
| Procyanidins | Kd (mM) | n |
| B1 | 2.9 ± 1.4 | 2.6 ± 0.4 |
| B2 | 1.2 ± 0.4 | 3.6 ± 0.5 |
| B3 | 8.0 ± 0.9 | 3.3 ± 0.5 |
| B4 | 1.4 ± 0.4 | 3.7 ± 0.3 |
As shown by the standard deviation measures, values were obtained with a better accuracy than values previously reported [16]. This is probably due to a tidy control of peptide vs tannin concentration all along the titration. This accuracy is of crucial importance because it allows us to have sufficient confidence in Kd and n values and in their differences to rank tannins with respect to their affinity towards the PRP peptide.
3.2 Diffusion measurements
NMR diffusion experiments are powerful for detecting the association of small molecules with a host molecule not exceeding a few kDa in molecular weight. Indeed, in such a case it is easy to follow changes in the diffusion speed of the host all along the binding of guest molecules [24] for determining both Kd and the number of fixation sites. Thus, following changes of the 1.3 kDa PRP peptide diffusion rate with increased tannin content (dimer molecular weight: 479 Da) is possible as shown in Fig. 3. A progressive decrease of D is observed as the tannin content increases, in agreement with the increase of the overall complex size. Experimental data for each tannin were fitted using Eq. (2) (in this case, Aobs = Dobs) and allowed to measure Kd and n measurements. The values found for each tannin were used to calculate the averaged values reported on Table 1. D values evolve from approximately 3.0 10−10 m2.s−1 for the peptide alone to 2.0 10−10 m2.s−1 when complexed with procyanidin dimers. This corresponds to a change in the hydrodynamic radius rH from 6.8 for the peptide alone to 8.2 Å for the petide complexed with at least three procyanidin dimers.
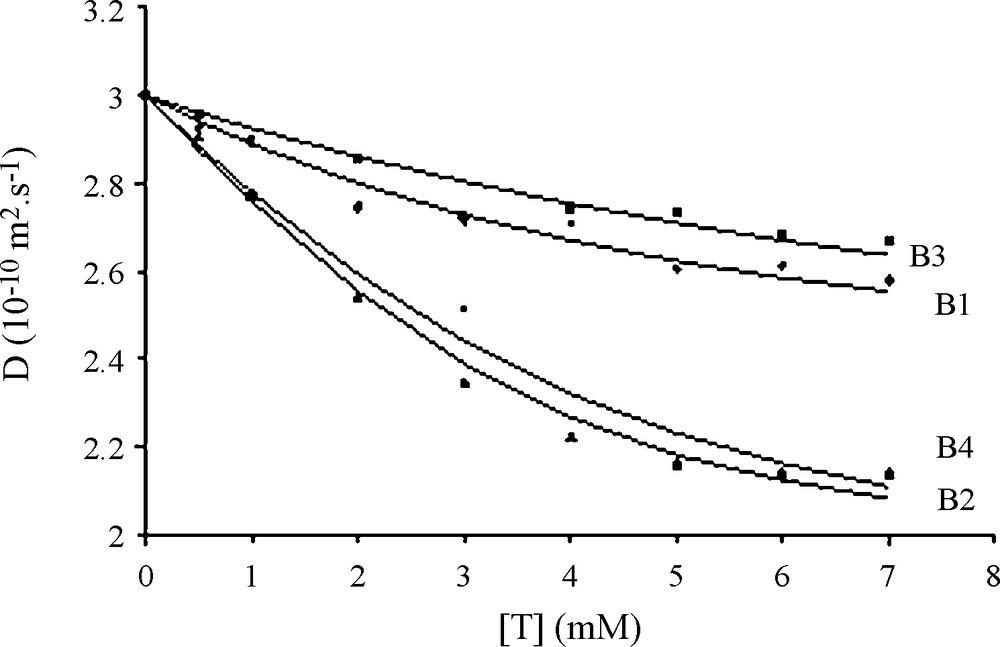
Diffusion coefficient evolution of 1 mM IB714 peptide (D, 10-10 m2 s-1) with increasing tannins concentrations. Diamond (B1) triangle (B2) square (B3) and circle (B4) are the experimental data; Solid lines represent the best fit using eq. (1) with Kd = 3.5 mM, n = 2.6, (Dmax = 0.75 and (2 = 2.2 10-2 for B1; Kd = 0.8 mM, n = 3.6, (Dmax = 1.1 and (2 = 0.8 10-2 for B2; Kd = 8.5 mM, n = 3.3, (Dmax = 0.9 and (2 = 0.9 10-2 for B3; Kd = 1.4 mM, n = 3.7, (Dmax = 1.1 and (2 = 4.2 10-2 for B4.
4 Discussion
In the present work, the interactions between four different procyanidin 4-8 dimers and the human saliva protein fragment IB714 were monitored by 1H NMR spectroscopy. Using both chemical shift and diffusion coefficient, titration information on the stoichiometry of the complex and dissociation constants were determined using a multisite model of Protein-ligand for which each ligand exhibits the same affinity toward the protein as previously described [16,21]. Monitoring both chemical shift and diffusion variations gives rise to some crucial information. First, some information is obtained about the fixation sites: the number of fixation sites has been evaluated to three whatever the procyanidin dimer considered, and if the peptide protons that undergo the greater chemical shift variations can be assimilated to the fixation sites, three different sites can be identified: P2, P9-P10 and G13-G14. These sites are the same for each tannin. Second, some information is collected about tannin-saliva protein affinity. Dissociation constants (Kd) are obtained by fitting experimental data and give rise to value measurement with a good accuracy. These values vary from 1.2 to 8 mM, despite the tenuous chemical structure differences in between the four dimers studied. B2 and B4 dimers present a higher affinity than B1 and, a fortiori, B3 towards the saliva peptide IB7-14, leading to rank tannins as regard to their affinity. For these measurement, the procyanidin self-association process has been taken into account, the association constant of this phenomenon being closed to 7 M−1, whatever the procyanidin dimer considered [22]. Nevertheless, it is noteworthy that this phenomenon is favored at higher concentration than those we used: the critical micellar/aggregation concentration for dimers is above 17 mM, and titration experiments were done at lower procyanidin dimers concentrations (under 7 mM).
Of interest is to compare these affinity differences to 3D-structure of procyanidin dimers. 4-8 procyanidin dimers are known to exist upon two rotameric forms: a compact form, which is largely dominant for B1 and B3 (95%) and an extended form which exist, at a level up to 24% for B4 and, up to 45% for B2 [25]. This let us suggest that the extended conformation must facilitate the approach or/and the fixation of the tannin to the peptide: the more the extended conformation is preferred, the higher the affinity towards the PRP peptide is.
Acknowledgements
This work has been supported by the Conseil interprofessionnel de Bordeaux (CIVB), the CNRS, the université de Bordeaux 1. No conflict of interest exists for all the co-authors.


